Lysosome: regulator of lipid degradation pathways - PubMed (original) (raw)
Review
Lysosome: regulator of lipid degradation pathways
Carmine Settembre et al. Trends Cell Biol. 2014 Dec.
Abstract
Autophagy is a catabolic pathway that has a fundamental role in the adaptation to fasting and primarily relies on the activity of the endolysosomal system, to which the autophagosome targets substrates for degradation. Recent studies have revealed that the lysosomal-autophagic pathway plays an important part in the early steps of lipid degradation. In this review, we discuss the transcriptional mechanisms underlying co-regulation between lysosome, autophagy, and other steps of lipid catabolism, including the activity of nutrient-sensitive transcription factors (TFs) and of members of the nuclear receptor family. In addition, we discuss how the lysosome acts as a metabolic sensor and orchestrates the transcriptional response to fasting.
Keywords: Autophagy; FOXOs; TFEB; TP53; lipophagy; lysosome; mTORC1; nuclear receptors; transcription factors.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
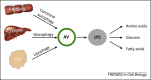
Figure 1
Autophagy mediates substrate catabolism during fasting. During nutrient deprivation proteins, glycogen and fat are sequestered by autophagosomes and targeted to lysosomes where they are degraded by resident hydrolases and transformed into amino acids, fatty acids, and glucose, which are then released into the cytoplasm to support cellular energetic demands. Abbreviations: AV, autophagosome; LYS, lysosome.
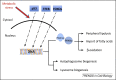
Figure 2
Proposed model of coordinated transcriptional regulation of lipid catabolism via forkhead box protein class O (FOXO), transcription factor EB (TFEB), p53, and nuclear receptors. The nuclear translocation of TFEB, p53, and FOXOs is induced under conditions of metabolic stress such as nutrient depletion and growth factor deprivation. These transcription factors (TFs) regulate directly the expression of autophagy genes and the nuclear receptor and co-receptor peroxisome proliferator-activated receptor (PPAR)α and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α), respectively, which control lipid catabolism.
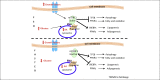
Figure 3
The lysosome as a regulator of lipid metabolism. In the fed state (upper panel), the presence of lysosomal amino acids (AA), glucose, and growth factors induces activation of mechanistic target of rapamycin complex 1 (mTORC1) on the lysosomal membrane. Active mTORC1 transcriptionally induces lipogenesis and adipogenesis by activating SREBPs and peroxisome proliferator-activated receptor (PPAR)γ transcription factors, respectively. Concomitantly, mTORC1 blocks autophagy and fatty acid oxidation via transcription factor EB (TFEB) and PPARα inhibition, respectively. In the fasted state (lower panel), lower levels of lysosomal AA, glucose, and growth factors induce mTORC1 detachment from the lysosomal membrane and its consequent inhibition. mTORC1 inhibition in turn activates TFEB and PPARα transcription factors, which leads to the transcriptional induction of lipophagy and β-oxidation, respectively. Concomitantly, SREBPs and PPARγ are no longer activated by mTORC1, therefore lipogenesis and adipogenesis are inhibited. Abbreviations: Rheb, Ras homolog enriched in brain; Rags, Ras-related small GTP-binding protein; SREBPs, sterol regulatory element-binding proteins.
Similar articles
- Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion.
Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. Zhou J, et al. Cell Res. 2013 Apr;23(4):508-23. doi: 10.1038/cr.2013.11. Epub 2013 Jan 22. Cell Res. 2013. PMID: 23337583 Free PMC article. - At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy.
Shen HM, Mizushima N. Shen HM, et al. Trends Biochem Sci. 2014 Feb;39(2):61-71. doi: 10.1016/j.tibs.2013.12.001. Epub 2013 Dec 24. Trends Biochem Sci. 2014. PMID: 24369758 Review. - Endocytosis and autophagy: Shared machinery for degradation.
Lamb CA, Dooley HC, Tooze SA. Lamb CA, et al. Bioessays. 2013 Jan;35(1):34-45. doi: 10.1002/bies.201200130. Epub 2012 Nov 13. Bioessays. 2013. PMID: 23147242 Review. - Dual suppressive effect of MTORC1 on autophagy: tame the dragon by shackling both the head and the tail.
Zhou J, Tan SH, Codogno P, Shen HM. Zhou J, et al. Autophagy. 2013 May;9(5):803-5. doi: 10.4161/auto.23965. Epub 2013 Feb 25. Autophagy. 2013. PMID: 23439250 Free PMC article. - Autophagy, lipophagy and lysosomal lipid storage disorders.
Ward C, Martinez-Lopez N, Otten EG, Carroll B, Maetzel D, Singh R, Sarkar S, Korolchuk VI. Ward C, et al. Biochim Biophys Acta. 2016 Apr;1861(4):269-84. doi: 10.1016/j.bbalip.2016.01.006. Epub 2016 Jan 14. Biochim Biophys Acta. 2016. PMID: 26778751 Review.
Cited by
- Lack of the Lysosomal Membrane Protein, GLMP, in Mice Results in Metabolic Dysregulation in Liver.
Kong XY, Kase ET, Herskedal A, Schjalm C, Damme M, Nesset CK, Thoresen GH, Rustan AC, Eskild W. Kong XY, et al. PLoS One. 2015 Jun 5;10(6):e0129402. doi: 10.1371/journal.pone.0129402. eCollection 2015. PLoS One. 2015. PMID: 26047317 Free PMC article. - Dietary cholesterol and apolipoprotein A-I are trafficked in endosomes and lysosomes in the live zebrafish intestine.
Otis JP, Shen MC, Caldwell BA, Reyes Gaido OE, Farber SA. Otis JP, et al. Am J Physiol Gastrointest Liver Physiol. 2019 Mar 1;316(3):G350-G365. doi: 10.1152/ajpgi.00080.2018. Epub 2019 Jan 10. Am J Physiol Gastrointest Liver Physiol. 2019. PMID: 30629468 Free PMC article. - Immunometabolic rewiring of tubular epithelial cells in kidney disease.
van der Rijt S, Leemans JC, Florquin S, Houtkooper RH, Tammaro A. van der Rijt S, et al. Nat Rev Nephrol. 2022 Sep;18(9):588-603. doi: 10.1038/s41581-022-00592-x. Epub 2022 Jul 7. Nat Rev Nephrol. 2022. PMID: 35798902 Review. - Role of lipid transfer proteins in loading CD1 antigen-presenting molecules.
Teyton L. Teyton L. J Lipid Res. 2018 Aug;59(8):1367-1373. doi: 10.1194/jlr.R083212. Epub 2018 Mar 19. J Lipid Res. 2018. PMID: 29559523 Free PMC article. Review. - Nutrient-sensing nuclear receptors coordinate autophagy.
Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Lee JM, et al. Nature. 2014 Dec 4;516(7529):112-5. doi: 10.1038/nature13961. Epub 2014 Nov 12. Nature. 2014. PMID: 25383539 Free PMC article.
References
- Wang T. The comparative physiology of food deprivation: from feast to famine. Annu. Rev. Physiol. 2006;68:223–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS078072/NS/NINDS NIH HHS/United States
- U54 HD083092/HD/NICHD NIH HHS/United States
- 250154/ERC_/European Research Council/International
- TCP12008/TI_/Telethon/Italy
- R01-NS078072/NS/NINDS NIH HHS/United States
- TGM11CB6/TI_/Telethon/Italy
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous