Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus - PubMed (original) (raw)
Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus
Ukpong B Eyo et al. J Neurosci. 2014.
Abstract
Microglia are highly dynamic immune cells of the CNS and their dynamism is proposed to be regulated by neuronal activities. However, the mechanisms underlying neuronal regulation of microglial dynamism have not been determined. Here, we found an increased number of microglial primary processes in the hippocampus during KA-induced seizure activity. Consistently, global glutamate induced robust microglial process extension toward neurons in both brain slices and in the intact brain in vivo. The mechanism of the glutamate-induced microglial process extension involves the activation of neuronal NMDA receptors, calcium influx, subsequent ATP release, and microglial response through P2Y12 receptors. Seizure-induced increases in microglial process numbers were also dependent on NMDA receptor activation. Finally, we found that P2Y12 KO mice exhibited reduced seizure-induced increases in microglial process numbers and worsened KA-induced seizure behaviors. Our results elucidate the molecular mechanisms underlying microglia-neuron communication that may be potentially neuroprotective in the epileptic brain.
Keywords: NMDA receptor; P2Y12 receptor; epilepsy; glutamate; microglia; process extension.
Copyright © 2014 the authors 0270-6474/14/3410528-13$15.00/0.
Figures
Figure 1.
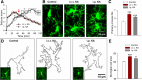
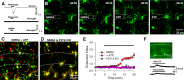
KA-induced seizures alter microglial morphology in vivo. A, Seizure scores following intraperitoneal and intracerebroventricular injections of KA identifying the period in which microglial morphology was studied (red arrow). B, Representative images of hippocampal microglia from control and KA-treated animals. C, Quantitative data showing KA increases primary microglial branch numbers. D, Outline of microglial morphologies in representative slices from control and KA-treated mice used to determine microglial area. E, Quantitative normalized area showing KA increases microglial cell area; *p < 0.05, ***p < 0.001.
Figure 2.
Glutamate induces microglial extension independent of action potential firing in slices and in vivo. A, Representative two-photon images from time-lapse movies at the beginning (i) and 15 min (ii) of glutamate (1 m
m
) showing that glutamate induces microglial process extension in the CA1, which is merged (iii) as a color-coded image of microglial morphology at the beginning of glutamate application (red) and after 15 min of glutamate exposure (green). Extending processes are obvious (shown in green) following glutamate application. B, Merged images from a time-lapse movie showing that TTX does not block glutamate-induced microglial process extension. C, High K+ does not induce microglial process extension. D, Representative electrophysiological tracings showing that glutamate-induced action potentials are blocked by TTX (1 μ
m
). E, Quantitative summary of microglial process extension through time under control media change, high K+, glutamate, and glutamate with TTX application from several experiments. F, Average extension index at different concentrations of glutamate (note that n = only 3 of 10 slices at 0.5 m
m
as there were no responses in the remaining 7 slices); ***p < 0.001. G, Images from a time-lapse sequence of Sytox-labeled slices. Neurons are healthy and unlabeled during 15 min of glutamate (1 m
m
) application. The few labeled cells are crescent shaped like endothelial cells (arrow). Neurons take up Sytox after a laser burn-induced injury (asterisk). H–J, Microglial processes extend during glutamate application in cortical slices (1 m
m
; H, I) as well as in vivo (5 m
m
; J) with bulbous ending (J, arrowheads).
Figure 3.
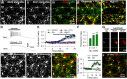
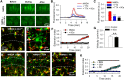
Glutamate increases microglial contact with neurons. A, Left, Larger field of view showing neurons and microglia in CA1 in GFP-YFP double transgenic mice. Right, The boxed region in the left image shows that during and after glutamate application microglial processes make contact with neuronal dendrites in the SP. Points of contact are depicted with red arrowheads. B, A _z_-stack projection of a representative hippocampal slice showing florescent microglia (green) and neurons (red) in GFP-YFP double transgenic mice. C, Time-lapse images of the boxed region in B show microglial contact with neuronal somata. Points of contact are depicted with white arrows. D, Single plane images of neurons and microglia in C at 2 μm intervals show contact (white arrows) in the same plane. E, Quantitative data show that glutamate (1 m
m
) application increases microglial contact with neuronal somata. F, Merge color-coded images from time-lapse movies of microglial process extension during glutamate application in the presence of astroglial toxin, FAC (1 m
m
). G, Quantitative summary of microglial process extension during glutamate application with or without FAC (1 m
m
); n = 15 neurons in F and *p < 0.05.
Figure 4.
Glutamate induces microglial process extension through NMDA receptors. A–C, Merged color-coded images from time-lapse movies showing that glutamate (1 m
m
) induces microglial extension (A) even in the presence of CNQX (10 μ
m
; B), but fails to do so in the presence of AP5 (C; 100 μ
m
). D, Quantitative summary of A–C. E–G, NMDA (30 μ
m
) induces microglial process extension (E), which is blocked by AP5 (100 μ
m
; G), while KA (100 μ
m
) fails to do so (F). H, Quantitative summary of E–G. I, Seizure scores during intracerebroventricular delivery of KA in the presence or absence of AP5. J–L, Qualitative images (J, K) and quantitative summary (L) of microglial morphology at 45 min of intracerebroventricular KA delivery with or without AP5. AP5 reduces seizure-induced microglial morphological changes; *p < 0.05.
Figure 5.
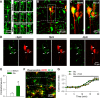
NMDA-induced microglial process extension requires purinergic signaling and P2Y12 receptor. A, CA1 neurons show large inward currents to puff application of glutamate (top trace) while microglia show no current responses to either glutamate (middle trace) or NMDA (bottom trace). B, Puff application of NMDA (100 μ
m
; 5 psi for 10 s) fails to induce microglial process extension while puff application of ATP (3 m
m
; 5 psi for 1 s) induces microglial process extension directly toward the pipette tip. C, D, NMDA (30 μ
m
) fails to induce robust microglial process extension in the presence of ATP (1 m
m
; C) or in P2Y12 KO tissues (D). E, Quantitative summary of the data presented in C and D. F, Top, Epifluorescence image of patched microglia in the CA1. Bottom, Representative tracing showing that bath application of ATP- and NMDA-induced outward currents in patched microglia in the CA1.
Figure 6.
NMDA-induced microglial process extension requires calcium influx and purine release from pannexin channels. A–C, Qualitative images of OGB-loaded hippocampal CA1 neurons before, during, and following a 2 min application of NMDA in normal ACSF (A, top) and in TTX containing nominally free calcium ACSF (A, bottom) with the quantitative summary in B and C. D, F, Representative image (D) and quantitative summary (F) showing that in the absence of extracellular calcium, microglial process extension to NMDA is abolished. E, Sample color-coded image of an ATP-induced chemotaxis at the beginning (00:00; red) and 30 min (00:30; green). Dashed lines indicate the location of the pipette. G, Quantitative data show similar extension velocities with or without extracellular calcium during ATP application through a pipette. H–J, Microglial process extension still occurs in the presence of P2X7 receptor antagonist BBG (10 μ
m
; H) and connexin channel blocker CBX (50 μ
m
; I) but not in the presence of pannexin channel blocker PB (5 m
m
; J). K, Quantitative summary of process extension under various conditions in H–J; ***p < 0.001. a.u., arbitrary units.
Figure 7.
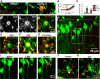
Injury-induced microglial process extension does not require NMDA receptors or calcium influx. A, Time-lapse images of microglia (green) during NMDA application (left) and following a laser-induced injury (asterisk, right) in the presence of propidium iodide (red). B, Propidium iodide uptake following laser-induced injury but not following glutamate/NMDA application. Arrow indicates time point of laser-induced injury. C, Number of propidium iodide-positive cells after application of glutamate, NMDA, and a corresponding laser burn. D, Laser-induced tissue injury induces robust microglial extension toward the injury site in control slices. E–G, Microglial processes still extend toward the injury site in the absence of extracellular calcium (E, 0Ca2+) and presence of AP5 (F, 100 μ
m
) but not in P2Y12 KO slices (D). H, YFP-labeled neurons (green) in the presence of Sytox (red) in the CA1 of the hippocampus. I, Neurons in the boxed region in H before, 1 min, and 10 min after single-cell ablation. The neuron to be ablated is identified with a white asterisk. Subsequent to laser ablation, Sytox (red; white arrow) is taken up by the single neuron. J, K, Representative merged color-coded images of microglial processes before (red) and 15 min after (green) single neuron ablation under control (J) and in the presence of AP5 (100 μ
m
; K). The ablation site is identified with a white asterisk; ***p < 0.001.
Figure 8.
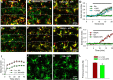
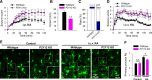
P2Y12 KO mice exhibit exacerbated seizure phenotypes and reduced microglial process numbers. A, Acute seizure scores in wild-type and P2Y12 KO mice following intraperitoneal injection of KA. B, Timing to seizure onset (stage 3 on the Racine scale) in wild-type and P2Y12 KO mice following intraperitoneal KA injection. C, Animal survival/ mortality rates in wild-type and P2Y12 KO mice following intraperitoneal KA injection. D, Acute seizure scores in wild-type and P2Y12 KO mice following intracerebroventricular injection of KA. E, F, Representative images of hippocampal microglia in the CA1 from wild-type and P2Y12 KO tissues under control and 45 min after intracerebroventricular KA conditions (E) and the corresponding quantitative data of microglial primary process numbers (F); *p < 0.05, **p < 0.01.
Figure 9.
Schematic model for glutamate-induced microglial process extension in epilepsy. During periods of intense neuronal hyperactivity as occurs in epilepsy, neurons release glutamate from presynaptic terminals that can activate AMPA receptors (AMPA-R) and NMDA receptors (NMDA-R) on postsynaptic sites. The NMDA-R activation leads to influx of extracellular calcium and such elevations of cytosolic calcium, through currently unknown mechanisms, result in the release of ATP possibly through either ion channels like pannexin 1(Px1) or prepackaged vesicles. Released ATP diffuses into the extracellular space forming a chemotactic gradient that activates microglial P2Y12 receptors (P2Y12-R) to induce microglial process extension toward neuronal elements.
Similar articles
- Activation of neuronal NMDA receptors triggers transient ATP-mediated microglial process outgrowth.
Dissing-Olesen L, LeDue JM, Rungta RL, Hefendehl JK, Choi HB, MacVicar BA. Dissing-Olesen L, et al. J Neurosci. 2014 Aug 6;34(32):10511-27. doi: 10.1523/JNEUROSCI.0405-14.2014. J Neurosci. 2014. PMID: 25100586 Free PMC article. - Status epilepticus induces a particular microglial activation state characterized by enhanced purinergic signaling.
Avignone E, Ulmann L, Levavasseur F, Rassendren F, Audinat E. Avignone E, et al. J Neurosci. 2008 Sep 10;28(37):9133-44. doi: 10.1523/JNEUROSCI.1820-08.2008. J Neurosci. 2008. PMID: 18784294 Free PMC article. - Distinct P2Y Receptors Mediate Extension and Retraction of Microglial Processes in Epileptic and Peritumoral Human Tissue.
Milior G, Morin-Brureau M, Chali F, Le Duigou C, Savary E, Huberfeld G, Rouach N, Pallud J, Capelle L, Navarro V, Mathon B, Clemenceau S, Miles R. Milior G, et al. J Neurosci. 2020 Feb 12;40(7):1373-1388. doi: 10.1523/JNEUROSCI.0218-19.2019. Epub 2020 Jan 2. J Neurosci. 2020. PMID: 31896671 Free PMC article. - Microglial circadian clock regulation of microglial structural complexity, dendritic spine density and inflammatory response.
Nakanishi H, Ni J, Nonaka S, Hayashi Y. Nakanishi H, et al. Neurochem Int. 2021 Jan;142:104905. doi: 10.1016/j.neuint.2020.104905. Epub 2020 Nov 18. Neurochem Int. 2021. PMID: 33217515 Review. - G-protein coupled purinergic P2Y12 receptor interacts and internalizes TauRD-mediated by membrane-associated actin cytoskeleton remodeling in microglia.
Chidambaram H, Das R, Chinnathambi S. Chidambaram H, et al. Eur J Cell Biol. 2022 Apr;101(2):151201. doi: 10.1016/j.ejcb.2022.151201. Epub 2022 Jan 25. Eur J Cell Biol. 2022. PMID: 35101770 Review.
Cited by
- Spatiotemporal profile of Map2 and microglial changes in the hippocampal CA1 region following pilocarpine-induced status epilepticus.
Schartz ND, Herr SA, Madsen L, Butts SJ, Torres C, Mendez LB, Brewster AL. Schartz ND, et al. Sci Rep. 2016 May 4;6:24988. doi: 10.1038/srep24988. Sci Rep. 2016. PMID: 27143585 Free PMC article. - Neuroinflammation and status epilepticus: a narrative review unraveling a complex interplay.
Foiadelli T, Santangelo A, Costagliola G, Costa E, Scacciati M, Riva A, Volpedo G, Smaldone M, Bonuccelli A, Clemente AM, Ferretti A, Savasta S, Striano P, Orsini A. Foiadelli T, et al. Front Pediatr. 2023 Nov 21;11:1251914. doi: 10.3389/fped.2023.1251914. eCollection 2023. Front Pediatr. 2023. PMID: 38078329 Free PMC article. Review. - Brake Early: RGS14 in CA2 Limits Seizures and Oxidative Stress After SE.
Metcalf CS. Metcalf CS. Epilepsy Curr. 2023 Sep 28;23(6):372-374. doi: 10.1177/15357597231199343. eCollection 2023 Nov-Dec. Epilepsy Curr. 2023. PMID: 38269350 Free PMC article. - Role of P2X7 Receptors in Immune Responses During Neurodegeneration.
Oliveira-Giacomelli Á, Petiz LL, Andrejew R, Turrini N, Silva JB, Sack U, Ulrich H. Oliveira-Giacomelli Á, et al. Front Cell Neurosci. 2021 May 26;15:662935. doi: 10.3389/fncel.2021.662935. eCollection 2021. Front Cell Neurosci. 2021. PMID: 34122013 Free PMC article. Review. - Microglia maintain the normal structure and function of the hippocampal astrocyte network.
Du Y, Brennan FH, Popovich PG, Zhou M. Du Y, et al. Glia. 2022 Jul;70(7):1359-1379. doi: 10.1002/glia.24179. Epub 2022 Apr 8. Glia. 2022. PMID: 35394085 Free PMC article.
References
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials