Identification of cleavage sites leading to the shed form of the anti-aging protein klotho - PubMed (original) (raw)
. 2014 Sep 2;53(34):5579-87.
doi: 10.1021/bi500409n. Epub 2014 Aug 20.
Affiliations
- PMID: 25110992
- PMCID: PMC4151695
- DOI: 10.1021/bi500409n
Identification of cleavage sites leading to the shed form of the anti-aging protein klotho
Ci-Di Chen et al. Biochemistry. 2014.
Abstract
Membrane protein shedding is a critical step in many normal and pathological processes. The anti-aging protein klotho (KL), mainly expressed in kidney and brain, is secreted into the serum and CSF, respectively. KL is proteolytically released, or shed, from the cell surface by ADAM10 and ADAM17, which are the α-secretases that also cleave the amyloid precursor protein and other proteins. The transmembrane KL is a coreceptor with the FGF receptor for FGF23, whereas the shed form acts as a circulating hormone. However, the precise cleavage sites in KL are unknown. KL contains two major cleavage sites: one close to the juxtamembrane region and another between the KL1 and KL2 domains. We identified the cleavage site involved in KL release by mutating potential sheddase(s) recognition sequences and examining the production of the KL extracellular fragments in transfected COS-7 cells. Deletion of amino acids T958 and L959 results in a 50-60% reduction in KL shedding, and an additional P954E mutation results in further reduction of KL shedding by 70-80%. Deletion of amino acids 954-962 resulted in a 94% reduction in KL shedding. This mutant also had moderately decreased cell surface expression, yet had overall similar subcellular localization as that of WT KL, as demonstrated by immunofluorescence. Cleavage-resistant mutants could function as a FGFR coreceptor for FGF23, but they lost activity as a soluble form of KL in proliferation and transcriptional reporter assays. Cleavage between the KL1 and KL2 domains is dependent on juxtamembrane cleavage. Our results shed light onto mechanisms underlying KL release from the cell membrane and provide a target for potential pharmacologic interventions aimed at regulating KL secretion.
Figures
Figure 1
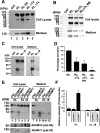
ADAM10 and ADAM17 cleavage site motif prediction in KL protein. (A) Schematic diagram of the KL protein structure and its processing in COS-7 cells. KL protein contains a signal sequence (SS), two homologous domains (KL1 and KL2), a transmembrane domain (TM), and a short cytoplasmic domain (CD). The anti-KL antibody recognition region and the length of the fragments are indicated. The predicted recognition sites are in bold. Dashes (−) in the amino acid sequence indicate cleavage positions. (B) A sequence alignment of 34 known substrate proteins compiled by Caescu et al. was used to generate a sequence logo with the program WebLogo 3.0. In the logo, the height of a particular letter is proportional to the log of its frequency in the sequence alignment.
Figure 2
Determination of the potential shedding sites and sheddases in KL in COS-7 cells. (A–C, E) Western blots from COS-7 cells transiently transfected with empty vector control (Ctrl) or with the KL plasmids as indicated. Forty-eight hours post-transfection, cells were incubated in serum-free media for 2 h, and the protein samples were collected from either the cell lysate (Cell lysate) or the medium (Medium). The medium samples in panel A were from 48 h post-transfection conditioned medium (CM), whereas in panels B and C, the serum-free medium was collected after 2 h incubation and TCA-precipitated as described in the Experimental Procedures. The estimated molecular weights of the KL fragments in the medium are indicated (130 and 70 kDa). (D) Statistical analysis of the results from panels A–C. The intensities of the 130 kDa bands were analyzed and normalized to that of the KL bands from the tissue lysate using the average intensity of the controls as 100% from 3 to 4 independent experiments. Error bar indicates standard deviation. Significance of results was determined using Student’s _t_-test: *, p < 0.05; **, p < 0.005. (E) Cotransfection experiments of KL WT and KL D9 with either ADAM10 or ADAM17. Anti-HA antibody detects HA-tagged ADAM10, whereas anti-V5 antibody detects V5-tagged ADAM17. (F) Statistical analysis of the results from panel E. The intensities of the 70 kDa bands in the medium were analyzed and normalized to the that of total KL bands from the tissue lysate using the average intensity of the controls as 100% from 3 independent experiments. Error bar indicates standard deviation. Significance of results was determined using Student’s _t_-test: *, p < 0.05.
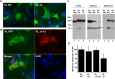
Figure 3
KLΔ9 mutant colocalization with KL WT and cell-surface distribution in COS-7 cells. (A) Indirect immunofluorescence showed subcellular localization pattern of KL WT and KLΔ9 mutant. COS-7 cells transfected with KL WT or KLΔ9 mutant for 48 h were fixed, and indirect immunofluorescence was performed using anti-V5 monoclonal antibody. Green: anti-V5 (Alexa 488); blue: DAPI nucleus staining. (B) Colocalization of KL GFP and KLΔ9 mutant with V5-tag. Green: GFP; red: anti-V5 (Alexa 594); blue: DAPI. (C) Western blots from COS-7 cells transiently transfected with the KL plasmids as indicated. Forty-eight hours post-transfection, the conditioned medium (Medium) was collected. The cells were surface-labeled with biotin, and the protein samples were collected from the cell lysates (Cell lysate). The biotinylated samples (Membrane) were pulled down by neutravidin beads. (D) Statistical analysis of the results from panel C. The intensities of the 130 kDa bands from the cell surface were analyzed and normalized to that of the total KL bands from the tissue lysate using the average intensity of the controls as 100% from 3 independent experiments. The arrowhead in the middle panel in C indicates nonspecific bands (arrowhead) at the 70 kDa position in the CM samples, likely from the interaction of the antibody with serum albumin. Error bar indicates standard deviation. Significance of results was determined using Student’s _t_-test: *, p < 0.05.
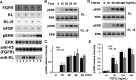
Figure 4
KLΔ9 mutant showed similar function as KL WT in downstream pERK signaling. HEK 293 cells transiently transfected with FGFR1c and either KL WT or KLΔ9 were treated with 10 ng/mL FGF23 for 30 min to activate FGFR1c signaling. (A) Representative western blot showing differences in ERK phosphorylation compared to total expression of ERK after transfection of KL WT or KLΔ9. Lanes 1 and 2 are negative controls without KL, and lanes 7 and 8 are positive controls with bFGF. The antibodies used are indicated. (B) Bar graph depicting no change in ERK phosphorylation normalized to total ERK expression (error bars are ±SEM; n = 3) when comparing KLΔ9 to KL WT. (C) Same experiments as in panel A with 0, 10, 20, 30, and 45 min time points for ERK phosphorylation kinetics analysis. (D) Statistical analysis of the results from panel C. The intensities of the pERK bands were normalized to that of the total ERK bands from 3 independent experiments. (E) FGF23 dose–response experiments. Similar experiments as those in panels A and C, with different doses of FGF23 for 10 min as indicated. (F) Statistical analysis of the results from panel E. Error bar indicates standard deviation. Significance of results was determined using Student’s _t_-test: *, p < 0.05.
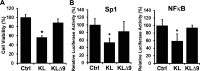
Figure 5
CM of COS-7 cells transfected with KLΔ9 has less inhibition of proliferation and transcription factor reporter activity in MO3.13 cells compared to the that from CM of KL WT. (A) MO3.13 cells were incubated with CM from either empty vector, KL WT, or KL Δ9 transfected COS-7 cells for 48 h and assayed for cell viability. Asterisks (*) indicate statistical significance of p < 0.01 by Student’s t test. Error bars indicate standard deviation. Results are from 3 independent experiments. (B) Luciferase assay of MO3.13 cells transfected with luciferase reporters as indicated. Cells were incubated with CM from either empty vector, KL WT, or KL Δ9 transfected COS-7 cells for 24 h and tested using the luciferase assay. The luminescent signals were normalized to Renilla luciferase. Luciferase activity was calculated relative to that of the control (CM from empty vector transfected COS-7 cells), which was given a value of 100%. Asterisks (*) indicate statistical significance of p < 0.01 by Student’s t test. Error bars indicate standard deviation.
Similar articles
- Identification of the cleavage sites leading to the shed forms of human and mouse anti-aging and cognition-enhancing protein Klotho.
Chen CD, Li Y, Chen AK, Rudy MA, Nasse JS, Zeldich E, Polanco TJ, Abraham CR. Chen CD, et al. PLoS One. 2020 Jan 13;15(1):e0226382. doi: 10.1371/journal.pone.0226382. eCollection 2020. PLoS One. 2020. PMID: 31929539 Free PMC article. - Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17.
Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Chen CD, et al. Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19796-801. doi: 10.1073/pnas.0709805104. Epub 2007 Dec 3. Proc Natl Acad Sci U S A. 2007. PMID: 18056631 Free PMC article. - Shedding of klotho by ADAMs in the kidney.
van Loon EP, Pulskens WP, van der Hagen EA, Lavrijsen M, Vervloet MG, van Goor H, Bindels RJ, Hoenderop JG. van Loon EP, et al. Am J Physiol Renal Physiol. 2015 Aug 15;309(4):F359-68. doi: 10.1152/ajprenal.00240.2014. Epub 2015 Jul 8. Am J Physiol Renal Physiol. 2015. PMID: 26155844 - [Discovery of alpha-Klotho and FGF23 unveiled new insight into calcium and phosphate homeostasis].
Nabeshima Y. Nabeshima Y. Clin Calcium. 2008 Jul;18(7):923-34. Clin Calcium. 2008. PMID: 18591743 Review. Japanese. - α-Klotho's effects on mineral homeostasis are fibroblast growth factor-23 dependent.
Erben RG. Erben RG. Curr Opin Nephrol Hypertens. 2018 Jul;27(4):229-235. doi: 10.1097/MNH.0000000000000415. Curr Opin Nephrol Hypertens. 2018. PMID: 29851418 Free PMC article. Review.
Cited by
- Klotho in Clinical Nephrology: Diagnostic and Therapeutic Implications.
Neyra JA, Hu MC, Moe OW. Neyra JA, et al. Clin J Am Soc Nephrol. 2020 Dec 31;16(1):162-176. doi: 10.2215/CJN.02840320. Epub 2020 Jul 22. Clin J Am Soc Nephrol. 2020. PMID: 32699047 Free PMC article. Review. - Klotho, the Key to Healthy Brain Aging?
Vo HT, Laszczyk AM, King GD. Vo HT, et al. Brain Plast. 2018 Aug 10;3(2):183-194. doi: 10.3233/BPL-170057. Brain Plast. 2018. PMID: 30151342 Free PMC article. Review. - Klotho's impact on diabetic nephropathy and its emerging connection to diabetic retinopathy.
Tang A, Zhang Y, Wu L, Lin Y, Lv L, Zhao L, Xu B, Huang Y, Li M. Tang A, et al. Front Endocrinol (Lausanne). 2023 Apr 18;14:1180169. doi: 10.3389/fendo.2023.1180169. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37143722 Free PMC article. Review. - Relationship between klotho and physical function in healthy aging.
Arroyo E, Leber CA, Burney HN, Narayanan G, Moorthi R, Avin KG, Warden SJ, Moe SM, Lim K. Arroyo E, et al. Sci Rep. 2023 Nov 30;13(1):21158. doi: 10.1038/s41598-023-47791-5. Sci Rep. 2023. PMID: 38036596 Free PMC article. - Role of Klotho Protein in Neuropsychiatric Disorders: A Narrative Review.
Birdi A, Tomo S, Yadav D, Sharma P, Nebhinani N, Mitra P, Banerjee M, Purohit P. Birdi A, et al. Indian J Clin Biochem. 2023 Jan;38(1):13-21. doi: 10.1007/s12291-022-01078-0. Epub 2022 Aug 13. Indian J Clin Biochem. 2023. PMID: 36684492 Free PMC article. Review.
References
- Kuro-o M.; Matsumura Y.; Aizawa H.; Kawaguchi H.; Suga T.; Utsugi T.; Ohyama Y.; Kurabayashi M.; Kaname T.; Kume E.; Iwasaki H.; Iida A.; Shiraki-Iida T.; Nishikawa S.; Nagai R.; Nabeshima Y. I. (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51. - PubMed
- Duce J. A.; Podvin S.; Hollander W.; Kipling D.; Rosene D. L.; Abraham C. R. (2008) Gene profile analysis implicates klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia 56, 106–117. - PubMed
- Kurosu H.; Yamamoto M.; Clark J. D.; Pastor J. V.; Nandi A.; Gurnani P.; McGuinness O. P.; Chikuda H.; Yamaguchi M.; Kawaguchi H.; Shimomura I.; Takayama Y.; Herz J.; Kahn C. R.; Rosenblatt K. P.; Kuro-o M. (2005) Suppression of aging in mice by the hormone klotho. Science 309, 1829–1833. - PMC - PubMed
- Chen C. D.; Sloane J. A.; Li H.; Aytan N.; Giannaris E. L.; Zeldich E.; Hinman J. D.; Dedeoglu A.; Rosene D. L.; Bansal R.; Luebke J. I.; Kuro-o M.; Abraham C. R. (2013) The antiaging protein klotho enhances oligodendrocyte maturation and myelination of the CNS. J. Neurosci. 33, 1927–1939. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous