DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome - PubMed (original) (raw)
DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome
Alessandro Costa et al. Elife. 2014.
Abstract
The Cdc45/Mcm2-7/GINS (CMG) helicase separates DNA strands during replication in eukaryotes. How the CMG is assembled and engages DNA substrates remains unclear. Using electron microscopy, we have determined the structure of the CMG in the presence of ATPγS and a DNA duplex bearing a 3' single-stranded tail. The structure shows that the MCM subunits of the CMG bind preferentially to single-stranded DNA, establishes the polarity by which DNA enters into the Mcm2-7 pore, and explains how Cdc45 helps prevent DNA from dissociating from the helicase. The Mcm2-7 subcomplex forms a cracked-ring, right-handed spiral when DNA and nucleotide are bound, revealing unexpected congruencies between the CMG and both bacterial DnaB helicases and the AAA+ motor of the eukaryotic proteasome. The existence of a subpopulation of dimeric CMGs establishes the subunit register of Mcm2-7 double hexamers and together with the spiral form highlights how Mcm2-7 transitions through different conformational and assembly states as it matures into a functional helicase.
Keywords: AAA+ ATPase; DNA replication; Mcm2-7; helicase; motor proteins; replication fork.
Copyright © 2014, Costa et al.
Conflict of interest statement
MRB: Reviewing editor, eLife.
The other authors declare that no competing interests exist.
Figures
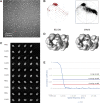
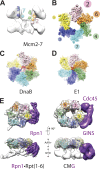
Figure 1.. 18 Å resolution of a CMG–DNA–ATPγS complex.
(A) Top-down view (N-terminal MCM face) of the CMG highlighting subunit positions. (B) Docking of homology models into the assembly. (C) Docked structures into segmented density for: _top_—a near-full-length, archaeal MCM monomer Mcm4 (PDB ID 3F9V); _middle_—the GINS complex (PDB ID 2Q9Q and 3ANW, ‘Materials and methods’); _bottom_—the archaeal Mcm N-terminal domain hexamer (PDB ID 1LTL and 2VL6, ‘Materials and methods’). (D) The N- and C-terminal domains of Mcm2-7 (colors) differentially flex around the helicase ring, with GINS–Cdc45 (white) wedging open Mcm5 in particular. (E) The N-terminal domains of Mcm2-7 are relatively planar, and are fit best by a hexameric, DNA-free structure of the archaeal MCM NTDs, indicating the observed intra-subunit flexing derives from ATPase domain movement. DOI:
http://dx.doi.org/10.7554/eLife.03273.003
Figure 1—figure supplement 1.. Overview of EM data.
(A) Representative micrographs. (B) Comparison of Euler plots obtained with either RELION (left) or SPARX (right) three-dimensional structure refinement. (C) Comparison between two-dimensional projections of the SPARX-refined, three-dimensional structure and reference-free, two-dimensional class averages. (D) Comparison of the independently determined, three-dimensional structures of DNA- and ATPγS-bound CMG particles generated with RELION and SPARX. (E) Fourier-shell correlation showing the calculated resolution for the 3D EM models. DOI:
http://dx.doi.org/10.7554/eLife.03273.004
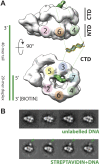
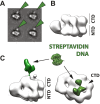
Figure 2.. Polarity of DNA binding by the CMG.
(A) Observed experimental density seen at low contours reveals a rod-shaped extension (green) of comparable length to that expected for a 20mer DNA duplex that extends from the Mcm C-terminal motor domain. This feature is absent in DNA-free CMG reconstructions (Costa et al., 2011). A schematic of the relative single- and double-stranded DNA regions of the substrate used for the present studies is shown at left. (B) Comparison of DNA–CMG class averages with and without streptavidin-labeling clearly marks the duplex end of the 3′-tailed duplex substrate. Note how the streptavidin density sits at a distance from the body of the CMG, indicating that the majority of the duplex region of the substrate is not bound by the CMG. DOI:
http://dx.doi.org/10.7554/eLife.03273.005
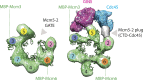
Figure 3.. Cdc45 is positioned to permit trapping of single-stranded DNA.
(A) _Top_—Segmented electron density corresponding to Cdc45. A prominent horseshoe-shaped region fits well to the catalytic core of the homologous RecJ exonuclease (PDB ID 1IR6). _Bottom_—Docked models of RecJ and full-length GINS (generated using an archaeal Psf1 homolog, PDB IDs 2Q9Q and 3ANW) into DNA-bound CMG reconstructions highlight a previously unobserved interaction between the B-domain of Psf1 and the exonuclease-like domain of Cdc45. (B) An extension of the Cdc45 RecJ-like region contacts and interdigitates between the A-domains of the Mcm2 and Mcm5 N-terminal regions. (C) The Mcm2-7 central channel (black line) and the Cdc45 DNA tracking groove (red arrow) are offset by ∼90°. (D) Schematic showing how the single-stranded DNA-binding groove of the Cdc45 RecJ-like domain could facilitate the capture of a leading strand segment if the Mcm5-2 DNA gate were to transiently open. DOI:
http://dx.doi.org/10.7554/eLife.03273.006
Figure 3—figure supplement 1.. Comparison between the open N-terminal domain of Drosophila Mcm2-7 and the closed N-terminal region of the CMG.
Docking of the Mcm2-7 DNA-interaction collar, GINS, and the RecJ exonuclease domain occupies most of the CMG density, with the exception of some unoccupied density shown in gray. We have tentatively assigned this region to the otherwise unaccounted for C-terminal domain of Cdc45; however, the density could also correspond to the disordered N-terminal tail of Mcm2, which may become structured upon Cdc45 binding. DOI:
http://dx.doi.org/10.7554/eLife.03273.007
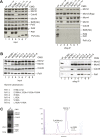
Figure 3—figure supplement 2.. The Psf1 C-terminus and Cdc45 N-terminus are critical for the CMG formation.
(A) (Left panel) Expression levels of individual subunits used for assessing the CMG formation were detected by antibodies against specific proteins as indicated (asterisk indicates a degradation product of Psf2). Loading was controlled by an α-tubulin antibody. (Right panel) Co-immunoprecipitation (IP) experiments analyzing CMG stability. Lanes are as follows: (1) wild-type CMG; (2 and 5–7) CMG with various C-terminal truncations in Psf1; (3) CMG without MCM5; (4) CMG without Cdc45. Immunoprecipitation of FLAG-tagged Mcm3 subunit pulls down intact CMG complexes (lane 1). By contrast, removal of the entire Psf1 B-domain (lane 2, Psf11−139), smaller truncations of Psf1 B-domain (lanes 5–7), the absence of Mcm5 (lane 3), or the absence of Cdc45 (lane 4) all disrupt the CMG formation. (B) (Left panel) Expression levels of individual subunits used for assessing the CMG formation were detected by antibodies against specific proteins as indicated. (Right panel) Co-immunoprecipitation (IP) experiments analyzing the CMG stability. Lanes are as follows: (1) wild-type CMG; (2) CMG expression without Mcm5; (3) CMG expression without Mcm3; (4–9) CMG expression with various alanine substitutions in Psf1 C-terminal helix. Immunoprecipitation of Mcm3 via a FLAG-tag on this subunit pulls down the intact CMG complex (lane 1), and most point mutations in the C-terminal helix of Psf1 do not appear to compromise the CMG formation (lanes 4 and 6–9). By contrast, the alanine substitutions in four residues in the B-domain of Psf1 (lane 5), the absence of Mcm5 (lane 2), or a lack of Mcm3 (the bait protein, lane 3) disrupt the CMG. The different alanine substitutions in Psf1 are listed below the gel at left. (C) Purification of the CMG with Cdc45Δ1−99. (Left) Input (4 μl of 30 ml-total clarified whole cell lysate) and IP (4 μl out of 5 ml-total eluate following anti-FLAG affinity purification) of the CMG component utilizing Cdc45Δ1−99 are shown. The FLAG-tag on Mcm3 was used as bait for the IP. Co-immunoprecipitated proteins were separated by SDS-PAGE (10%) and the CMG subunits were detected by antibodies against several CMG proteins as indicated on the right of the western blot. Mcm2 and Mcm5 represent the Mcm2-7 complex; Psf1 and Psf3 were used to follow the GINS complex. The experiment shows a near total loss of Psf1 and Psf3 from the CMG when the N-terminus of Cdc45 is truncated. (Right) MonoQ profile for the CMG purification, with the expected positions for various constituents labeled (FLAG-Mcm3 alone, Mcm2-7 and the intact CMG complex). The CMG formation was abrogated when the N-terminus of Cdc45 is removed. DOI:
http://dx.doi.org/10.7554/eLife.03273.008
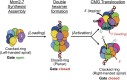
Figure 4.. Global comparison of the DNA-bound Mcm2-7 region of the CMG with other hexameric helicases and ATPases.
(A) Cut-away view (removing Mcm5) of the Mcm2-7 central channel highlights a spiral organization for the Mcm2-6-4-7-3 AAA+ ATPase regions. Colored spheres demarcate the approximate center of mass for AAA+ pore loops as derived from the docking of MCM AAA+ domain as shown in Figure 1B. (B) Top-down view (from the N-terminal face) of MCM AAA+ domains docked into the DNA-bound CMG reconstruction showing the existence of a right-handed spiral. The CMG density has been removed for clarity. (C) In the presence of a single-stranded DNA, bacterial DnaB can adopt a right-handed spiral with a moderately-wide pore (PDB ID 4ESV, Itsathitphaisarn et al., 2012). (D) The E1 helicase assembles into a right-handed spiral with a relatively narrow pore (PDB ID 2GXA, Enemark and Joshua-Tor, 2006). (E) Comparison of the AAA+ ring of the eukaryotic proteasome with Mcm2-7 region of the DNA- and ATPγS-bound CMG. The non-ATPase subunit Rpn1 binds to the side of the Rpt1-6 hetero-hexamer, wedging itself between the N-terminal and C-terminal tiers of the ATPase ring and helping to promote the formation of a right-handed ATPase domain spiral. Similar architectural features are apparent within the DNA–ATPγS–CMG complex, where GINS–Cdc45 occupy an analogous position. DOI:
http://dx.doi.org/10.7554/eLife.03273.009
Figure 5.. The CMG can form head-to-head dimers that establish the interactions between Mcm2-7 double hexamers.
(A) Experimentally observed 2D class averages showing that the CMG forms a defined double-hexamer in which the GINS-Cdc45 subcomplex is rotationally offset toward opposite sides of the Mcm2-7 rings. Arrowheads mark the position of GINS/Cdc45. The structure of the archaeal N-terminal double hexamer is shown to depict the six possible Mcm2-7 registers that were tested. The class averages show that the interface between the N-terminal collars can partially crack open in some instances, suggesting that this region is somewhat unstable when the CMG is bound to ssDNA. (B) Computationally derived 2D projections of double-hexameric 3D CMG models in which Mcm2-7 N-terminal domain dimers (modeled on an archaeal Mcm crystal structure of this region, PDB ID 1LTL) have been manually offset by distinct rotational registers. The reference-free class averages best resemble a model in which Mcm5 from ring 1 juxtaposes with Mcm4 from ring 2, although our data cannot formally rule out a configuration where Mcm4 might juxtapose with Mcm3 or Mcm2 (although in these configurations the two Mcm2/5 gates would still remain misaligned). Arrowheads mark the position of GINS/Cdc45**.** (C) Cartoon representation of the Mcm2-7 double ring register formed in head-to-head dimers of the CMG. DOI:
http://dx.doi.org/10.7554/eLife.03273.010
Figure 6.. Overview of Mcm2-7 organization, remodeling, and gate status during initiation and CMG formation.
Following synthesis and assembly, the isolated metazoan Mcm2-7 motor forms an inactive left-handed spiral, irrespective of nucleotide state, with a discontinuity between Mcm5 and Mcm2. The action of ORC, Cdc6 and Cdt1 results in the loading of a planar, head-to-head Mcm2-7 double hexamer onto a duplex DNA in which the Mcm2/5 gates are closed. Following loading, Dpb11-Sld2-Sld3 (11-2-3) chaperone GINS and Cdc45 onto the Mcm2-7 double hexamer and along with DDK promoted phosphorylation events, help promote both DNA melting, CMG formation and replication fork separation. Structural analysis of dimeric CMG particles (Figure 5) indicates that the Mcm2/5 gates are localized on the opposing sides of the dodecameric Mcm2-7 complex and that CMG formation precedes separation of the double hexamer. DOI:
http://dx.doi.org/10.7554/eLife.03273.011
Author response image 1.
Similar articles
- CryoEM structures of human CMG-ATPγS-DNA and CMG-AND-1 complexes.
Rzechorzek NJ, Hardwick SW, Jatikusumo VA, Chirgadze DY, Pellegrini L. Rzechorzek NJ, et al. Nucleic Acids Res. 2020 Jul 9;48(12):6980-6995. doi: 10.1093/nar/gkaa429. Nucleic Acids Res. 2020. PMID: 32453425 Free PMC article. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review. - Mechanism of replication origin melting nucleated by CMG helicase assembly.
Lewis JS, Gross MH, Sousa J, Henrikus SS, Greiwe JF, Nans A, Diffley JFX, Costa A. Lewis JS, et al. Nature. 2022 Jun;606(7916):1007-1014. doi: 10.1038/s41586-022-04829-4. Epub 2022 Jun 15. Nature. 2022. PMID: 35705812 Free PMC article. - Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, O'Donnell ME. Georgescu R, et al. Proc Natl Acad Sci U S A. 2017 Jan 31;114(5):E697-E706. doi: 10.1073/pnas.1620500114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096349 Free PMC article. - Archaeal MCM Proteins as an Analog for the Eukaryotic Mcm2-7 Helicase to Reveal Essential Features of Structure and Function.
Miller JM, Enemark EJ. Miller JM, et al. Archaea. 2015 Oct 11;2015:305497. doi: 10.1155/2015/305497. eCollection 2015. Archaea. 2015. PMID: 26539061 Free PMC article. Review.
Cited by
- Characterization of a Novel MMS-Sensitive Allele of Schizosaccharomyces pombe mcm4.
Ranatunga NS, Forsburg SL. Ranatunga NS, et al. G3 (Bethesda). 2016 Oct 13;6(10):3049-3063. doi: 10.1534/g3.116.033571. G3 (Bethesda). 2016. PMID: 27473316 Free PMC article. - Replication Stress and Consequential Instability of the Genome and Epigenome.
Tsegay PS, Lai Y, Liu Y. Tsegay PS, et al. Molecules. 2019 Oct 27;24(21):3870. doi: 10.3390/molecules24213870. Molecules. 2019. PMID: 31717862 Free PMC article. Review. - Mechanism of DNA unwinding by hexameric MCM8-9 in complex with HROB.
Acharya A, Bret H, Huang JW, Mütze M, Göse M, Kissling V, Seidel R, Ciccia A, Guérois R, Cejka P. Acharya A, et al. Res Sq [Preprint]. 2023 Jun 26:rs.3.rs-3054483. doi: 10.21203/rs.3.rs-3054483/v1. Res Sq. 2023. PMID: 37461676 Free PMC article. Updated. Preprint. - A reconstituted system reveals how activating and inhibitory interactions control DDK dependent assembly of the eukaryotic replicative helicase.
Herrera MC, Tognetti S, Riera A, Zech J, Clarke P, Fernández-Cid A, Speck C. Herrera MC, et al. Nucleic Acids Res. 2015 Dec 2;43(21):10238-50. doi: 10.1093/nar/gkv881. Epub 2015 Sep 3. Nucleic Acids Res. 2015. PMID: 26338774 Free PMC article. - Cdc45-induced loading of human RPA onto single-stranded DNA.
Szambowska A, Tessmer I, Prus P, Schlott B, Pospiech H, Grosse F. Szambowska A, et al. Nucleic Acids Res. 2017 Apr 7;45(6):3217-3230. doi: 10.1093/nar/gkw1364. Nucleic Acids Res. 2017. PMID: 28100698 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 15852/CRUK_/Cancer Research UK/United Kingdom
- R01 GM071747/GM/NIGMS NIH HHS/United States
- GM821972/GM/NIGMS NIH HHS/United States
- CA R37-30490/CA/NCI NIH HHS/United States
- GM071747/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous