Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics - PubMed (original) (raw)
Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics
Annalouise O'Connor et al. Mamm Genome. 2014 Dec.
Abstract
Intestinal microbial community structure is driven by host genetics in addition to environmental factors such as diet. In comparison with environmental influences, the effect of host genetics on intestinal microbiota, and how host-driven differences alter host metabolism is unclear. Additionally, the interaction between host genetics and diet, and the impact on the intestinal microbiome and possible down-stream effect on host metabolism is not fully understood, but represents another aspects of inter-individual variation in disease risk. The objectives of this study were to investigate how diet and genetic background shape microbial communities, and how these diet- and genetic-driven microbial differences relate to cardiometabolic phenotypes. To determine these effects, we used the 8 progenitor strains of the collaborative cross/diversity outbred mapping panels (C57BL/6J, A/J, NOD/ShiLtJ, NZO/HILtJ, WSB/EiJ, CAST/EiJ, PWK/PhJ, and 129S1/SvImJ). 16s rRNA profiling of enteric microbial communities in addition to the assessment of phenotypes central to cardiometabolic health was conducted under baseline nutritional conditions and in response to diets varying in atherogenic nutrient (fat, cholesterol, cholic acid) composition. These studies revealed strain-driven differences in enteric microbial communities which were retained with dietary intervention. Diet-strain interactions were seen for a core group of cardiometabolic-related microbial taxa. In conclusion, these studies highlight diet and genetically regulated cardiometabolic-related microbial taxa. Furthermore, we demonstrate the progenitor model is useful for nutrigenomic-based studies and screens seeking to investigate the interaction between genetic background and the phenotypic and microbial response to diet.
Figures
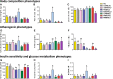
Fig. 1
Cardiometabolic phenotypes between inbred mouse strains fed a synthetic diet. Inbred mouse strains were purchased from Jackson Laboratories at ~6 weeks, and placed on purified synthetic diet (AIN-93). After 2 weeks, animals were weight, body composition assessed by MRI and fasting plasma samples collected. Differences in bodyweight (a), and body composition by MRI are depicted as percentage fat mass (b), percentage of lean mass (c). Fasting plasma levels of cholesterol (d), triglycerides (e) and TMAO (f) were quantitated. Measures of insulin sensitivity were assessed and included plasma glucose (g), insulin (h) and calculated HOMA-IR (i). _p_-value for ANOVA <0.05 for all phenotypes. Significant between-strain differences identified with Tukey’s HSD post hoc test. Strains not sharing letter are significantly different (p < 0.05)
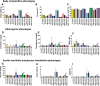
Fig. 2
Atherogenic diets induce strain-dependent differences in cardiometabolic phenotypes. Following 16 weeks on either a high-fat cholic acid (HFCA; open bars) diet or low-fat cholesterol-containing diet without cholic acid (LFCC; shaded bars). Differences in Bodyweight (a), and body composition by MRI are depicted as percentage fat mass (b), percentage of lean mass (c). Fasting plasma levels of cholesterol (d), triglycerides (e) and TMAO (f) were quantitated. Measures of insulin sensitivity were assessed and included plasma glucose (g), insulin (h) and calculated HOMA-IR (i). Significant within-strain differences between diet groups assessed by independent t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Fig. 3
Global regulation of intestinal microbiome communities by genetic background in mice fed a purified synthetic diet. (a) Principle coordinates analysis (PCoA) of unweighted uniFrac. (b) Principle component 1 of unweighted UniFrac. (c) Phyla level relative abundance data. (d) Linear discriminant analysis with effect size (LEfSe) identified differentially abundant taxa between mouse strains. Taxa enriched in A/J (yellow), C57Bl6/J (gray), 129S1/SvlmJ (pink), NOD/ShiLtJ (dark blue), NZO/HiLtJ (light blue), CAST/EiJ (green), PWK/PhJ (red), and WSB/EiJ (purple) meeting LDA significant threshold >2 are shown
Fig. 4
Global diet and strain-driven regulation of microbial beta-diversity. PCoA of Unweighted UniFrac of: (a) HFCA samples only; (b) LFCC samples only; (c) HFCA and LFCC samples combined; and, (d) HFCA, LFCC and AIN93M samples faceted by strain; (e) Linear discriminant analysis with Effect Size (LEfSe) identified differentially abundant taxa between HFCA and LFCC diet groups. Taxa enriched in HFCA diet (gray) and those enriched in LFCC diet (red) meeting LDA significant threshold >2 are shown. Taxa enriched in LF versus HFCA diet have a negative LDA score
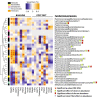
Fig. 5
Significant relationships between microbial taxa and cardiometabolic phenotypes exist in mice fed purified synthetic diets (baseline) and atherogenic diets for 16 weeks (post diet). Correlations between taxa and phenotype assessed by Spearman rho. Asterisks denotes significant relationship (p values adjusted FDR 10 %)
Similar articles
- Hepatic transcriptional profile reveals the role of diet and genetic backgrounds on metabolic traits in female progenitor strains of the Collaborative Cross.
Kim M, Huda MN, O'Connor A, Albright J, Durbin-Johnson B, Bennett BJ. Kim M, et al. Physiol Genomics. 2021 May 1;53(5):173-192. doi: 10.1152/physiolgenomics.00140.2020. Epub 2021 Apr 5. Physiol Genomics. 2021. PMID: 33818129 Free PMC article. - Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice.
Leist SR, Pilzner C, van den Brand JM, Dengler L, Geffers R, Kuiken T, Balling R, Kollmus H, Schughart K. Leist SR, et al. BMC Genomics. 2016 Feb 27;17:143. doi: 10.1186/s12864-016-2483-y. BMC Genomics. 2016. PMID: 26921172 Free PMC article. - Diet-induced obesity in genetically diverse collaborative cross mouse founder strains reveals diverse phenotype response and amelioration by quercetin treatment in 129S1/SvImJ, PWK/EiJ, CAST/PhJ, and WSB/EiJ mice.
Griffin LE, Essenmacher L, Racine KC, Iglesias-Carres L, Tessem JS, Smith SM, Neilson AP. Griffin LE, et al. J Nutr Biochem. 2021 Jan;87:108521. doi: 10.1016/j.jnutbio.2020.108521. Epub 2020 Oct 8. J Nutr Biochem. 2021. PMID: 33039581 - The Collaborative Cross mouse model for dissecting genetic susceptibility to infectious diseases.
Abu Toamih Atamni H, Nashef A, Iraqi FA. Abu Toamih Atamni H, et al. Mamm Genome. 2018 Aug;29(7-8):471-487. doi: 10.1007/s00335-018-9768-1. Epub 2018 Aug 24. Mamm Genome. 2018. PMID: 30143822 Review. - Foodomics as part of the host-microbiota-exposome interplay.
Putignani L, Dallapiccola B. Putignani L, et al. J Proteomics. 2016 Sep 16;147:3-20. doi: 10.1016/j.jprot.2016.04.033. Epub 2016 Apr 26. J Proteomics. 2016. PMID: 27130534 Review.
Cited by
- A comprehensive and comparative phenotypic analysis of the collaborative founder strains identifies new and known phenotypes.
Kollmus H, Fuchs H, Lengger C, Haselimashhadi H, Bogue MA, Östereicher MA, Horsch M, Adler T, Aguilar-Pimentel JA, Amarie OV, Becker L, Beckers J, Calzada-Wack J, Garrett L, Hans W, Hölter SM, Klein-Rodewald T, Maier H, Mayer-Kuckuk P, Miller G, Moreth K, Neff F, Rathkolb B, Rácz I, Rozman J, Spielmann N, Treise I, Busch D, Graw J, Klopstock T, Wolf E, Wurst W, Yildirim AÖ, Mason J, Torres A; Mouse Phenome Database Team; Balling R, Mehaan T, Gailus-Durner V, Schughart K, Hrabě de Angelis M. Kollmus H, et al. Mamm Genome. 2020 Feb;31(1-2):30-48. doi: 10.1007/s00335-020-09827-3. Epub 2020 Feb 14. Mamm Genome. 2020. PMID: 32060626 Free PMC article. - Dissecting the Genetic Architecture of Cystatin C in Diversity Outbred Mice.
Huda MN, VerHague M, Albright J, Smallwood T, Bell TA, Que E, Miller DR, Roshanravan B, Allayee H, Manuel de Villena FP, Bennett BJ. Huda MN, et al. G3 (Bethesda). 2020 Jul 7;10(7):2529-2541. doi: 10.1534/g3.120.401275. G3 (Bethesda). 2020. PMID: 32467129 Free PMC article. - Functional Genomics of Host-Microbiome Interactions in Humans.
Luca F, Kupfer SS, Knights D, Khoruts A, Blekhman R. Luca F, et al. Trends Genet. 2018 Jan;34(1):30-40. doi: 10.1016/j.tig.2017.10.001. Epub 2017 Oct 26. Trends Genet. 2018. PMID: 29107345 Free PMC article. Review. - Cross-species comparisons of host genetic associations with the microbiome.
Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE. Goodrich JK, et al. Science. 2016 Apr 29;352(6285):532-5. doi: 10.1126/science.aad9379. Science. 2016. PMID: 27126034 Free PMC article. Review. - Dual specificity phosphatase 6 deficiency is associated with impaired systemic glucose tolerance and reversible weight retardation in mice.
Pfuhlmann K, Pfluger PT, Schriever SC, Müller TD, Tschöp MH, Stemmer K. Pfuhlmann K, et al. PLoS One. 2017 Sep 5;12(9):e0183488. doi: 10.1371/journal.pone.0183488. eCollection 2017. PLoS One. 2017. PMID: 28873424 Free PMC article.
References
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. - DOI - PMC - PubMed
- Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. - DOI - PMC - PubMed
- Bennett BJ, Vallim TQdA, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-Oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. - DOI - PMC - PubMed
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous