Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage - PubMed (original) (raw)
Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage
Charlotte Frydenlund Michelsen et al. J Bacteriol. 2014 Nov.
Abstract
Interactions among members of polymicrobial infections or between pathogens and the commensal flora may determine disease outcomes. Pseudomonas aeruginosa and Staphylococcus aureus are important opportunistic human pathogens and are both part of the polymicrobial infection communities in human hosts. In this study, we analyzed the in vitro interaction between S. aureus and a collection of P. aeruginosa isolates representing different evolutionary steps of a dominant lineage, DK2, that have evolved through decades of growth in chronically infected patients. While the early adapted P. aeruginosa DK2 strains outcompeted S. aureus during coculture on agar plates, we found that later P. aeruginosa DK2 strains showed a commensal-like interaction, where S. aureus was not inhibited by P. aeruginosa and the growth activity of P. aeruginosa was enhanced in the presence of S. aureus. This effect is mediated by one or more extracellular S. aureus proteins greater than 10 kDa, which also suppressed P. aeruginosa autolysis and prevented killing by clinically relevant antibiotics through promoting small-colony variant (SCV) formation. The commensal interaction was abolished with S. aureus strains mutated in the agr quorum sensing system or in the SarA transcriptional virulence regulator, as well as with strains lacking the proteolytic subunit, ClpP, of the Clp protease. Our results show that during evolution of a dominant cystic fibrosis lineage of P. aeruginosa, a commensal interaction potential with S. aureus has developed.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
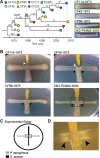
FIG 1
Overview of P. aeruginosa DK2 strains used in this study. (A) Tree showing the genetic relationship based on accumulations of single-nucleotide polymorphisms (SNPs) identified from genome sequencing (6, 7). The numbers in italics indicate the number of SNPs between the isolates. Symbols represent DK2 isolates sampled at different time points (indicated on the time line) from different patients with CF (indicated by symbol shape). On the right are colony morphologies of selected P. aeruginosa DK2 strains. (B) Cross-streak assay between selected P. aeruginosa DK2 stains and S. aureus strain JE2 cocultured on LB agar medium. White or black arrowheads indicate zones of bacterial inhibition or altered colony morphology (increased cell density), respectively. (C) Experimental setup of the cross-streak analysis between P. aeruginosa and S. aureus. The dashed square indicates the zone of interaction. (D) Zoom of interaction zone between P. aeruginosa DK2-P24M2-2003 and S. aureus JE2.
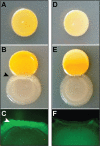
FIG 2
Monocultures of S. aureus JE2 WT (A) and the agrC mutant (D) or coculture with P. aeruginosa DK2-P24M2-2003/_gfp_AGA (B and E, respectively) by spot inoculation onto LB agar medium. The black arrowhead indicates altered P. aeruginosa colony morphology. The Gfp fluorescence signal of P. aeruginosa DK2-P24M2-2003/_gfp_AGA after 3 days of incubation is visualized in the zone of interaction with S. aureus JE2 WT (C) or the agrC mutant (F). The white arrowhead indicates increased Gfp expression by DK2-P24M2-2003/_gfp_AGA.
FIG 3
P. aeruginosa DK2-P24M2-2003 plated on top of LB agar plates without antibiotics (A) or with inhibitory levels of the antibiotics tobramycin (i.e., 15 μg/ml) (B), gentamicin (i.e., 38 μg/ml) (C), or ciprofloxacin (i.e., 3.5 μg/ml) (D). (A) Suppression of metallic sheen coverage of the P. aeruginosa DK2-P24M2-2003 lawn is observed in a zone surrounding the S. aureus JE2 WT culture supernatant (indicated by the arrowhead and scale bar) but not the agrC mutant supernatant. (B, C, and D) A halo of small colonies of P. aeruginosa DK2-P24M2-2003 is observed around the S. aureus JE2 WT culture supernatant (indicated by arrowheads) on antibiotic plates but not around the agrC mutant supernatant.
FIG 4
(A) Growth experiment with liquid cultures of P. aeruginosa DK2-P24M2-2003 treated with 10% unused TSB medium or control or S. aureus JE2 WT supernatant by measuring OD600 over time. (B, C, F, and G) The PI intensity histograms represent live/dead staining data from late-exponential-phase and stationary-phase DK2-P24M2-2003 control cultures (B and F, respectively) or from late-exponential-phase and stationary-phase cultures treated with S. aureus JE2 WT supernatant (C and G, respectively) as generated by flow cytometry. A distinct population of damaged/dead cells characterized by high PI uptake is evident only among cells from stationary-phase cultures treated with TSB (F). Miniature inserts display the TO-PI distribution of events from identical samples. Graphs represent sample data from a single culture representative of several independent experiments. (D and E) Control cultures of P. aeruginosa DK2-P24M2-2003 (D) but not cultures treated with S. aureus JE2 WT supernatant (E) show cell debris (indicated by the arrowhead) after 22 h of incubation.
FIG 5
(A) Colony morphologies of P. aeruginosa DK2-P24M2-2003 (left) and DK2-P24M2-TM1 (right) spotted (2 μl OD600 = 1) on top of LB agar medium. (B) Antibiotic resistance of P. aeruginosa DK2-P24M2-2003 (DK2-2003) and DK2-P24M2-TM1 (DK2-TM1) by determining the MIC using Etest strips of tobramycin (Tm), gentamicin (Gm), or ciprofloxacin (Ci).
Similar articles
- Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction.
Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, Ingmer H, Dorrestein PC, Jelsbak L. Frydenlund Michelsen C, et al. ISME J. 2016 Jun;10(6):1323-36. doi: 10.1038/ismej.2015.220. Epub 2015 Dec 18. ISME J. 2016. PMID: 26684729 Free PMC article. - Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model.
DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. DeLeon S, et al. Infect Immun. 2014 Nov;82(11):4718-28. doi: 10.1128/IAI.02198-14. Epub 2014 Aug 25. Infect Immun. 2014. PMID: 25156721 Free PMC article. - Subinhibitory Cefotaxime and Levofloxacin Concentrations Contribute to Selection of Pseudomonas aeruginosa in Coculture with Staphylococcus aureus.
Zhao K, Li J, Yang X, Zeng Q, Liu W, Wu Y, Zhou H, Prithiviraj B, Wang X, Zhou X, Chu Y. Zhao K, et al. Appl Environ Microbiol. 2022 Jun 28;88(12):e0059222. doi: 10.1128/aem.00592-22. Epub 2022 May 31. Appl Environ Microbiol. 2022. PMID: 35638844 Free PMC article. - Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus.
Yung DBY, Sircombe KJ, Pletzer D. Yung DBY, et al. Mol Microbiol. 2021 Jul;116(1):1-15. doi: 10.1111/mmi.14699. Epub 2021 Mar 8. Mol Microbiol. 2021. PMID: 33576132 Review. - Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction.
Magalhães AP, Jorge P, Pereira MO. Magalhães AP, et al. Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):712-728. doi: 10.1080/1040841X.2019.1700209. Epub 2019 Dec 13. Crit Rev Microbiol. 2019. PMID: 31835971 Review.
Cited by
- Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis.
Biswas L, Götz F. Biswas L, et al. Front Cell Infect Microbiol. 2022 Jan 6;11:824042. doi: 10.3389/fcimb.2021.824042. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35071057 Free PMC article. Review. - Pseudomonas aeruginosa surface motility and invasion into competing communities enhances interspecies antagonism.
Sánchez-Peña A, Winans JB, Nadell CD, Limoli DH. Sánchez-Peña A, et al. bioRxiv [Preprint]. 2024 Apr 4:2024.04.03.588010. doi: 10.1101/2024.04.03.588010. bioRxiv. 2024. PMID: 38617332 Free PMC article. Updated. Preprint. - Metabolic network modeling of microbial communities.
Biggs MB, Medlock GL, Kolling GL, Papin JA. Biggs MB, et al. Wiley Interdiscip Rev Syst Biol Med. 2015 Sep-Oct;7(5):317-34. doi: 10.1002/wsbm.1308. Epub 2015 Jun 24. Wiley Interdiscip Rev Syst Biol Med. 2015. PMID: 26109480 Free PMC article. Review. - Extracellular vesicles of Pseudomonas aeruginosa downregulate pyruvate fermentation enzymes and inhibit the initial growth of Staphylococcus aureus.
Ishiai T, Subsomwong P, Narita K, Kawai N, Teng W, Suzuki S, Sukchawalit R, Nakane A, Asano K. Ishiai T, et al. Curr Res Microb Sci. 2023 Apr 20;4:100190. doi: 10.1016/j.crmicr.2023.100190. eCollection 2023. Curr Res Microb Sci. 2023. PMID: 37131486 Free PMC article. - Iron Depletion Enhances Production of Antimicrobials by Pseudomonas aeruginosa.
Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. Nguyen AT, et al. J Bacteriol. 2015 Jul;197(14):2265-75. doi: 10.1128/JB.00072-15. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917911 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous