Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex - PubMed (original) (raw)
Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex
Caryn R Hale et al. Genes Dev. 2014.
Abstract
The effector complex of the Cmr/type III-B CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated) system cleaves RNAs recognized by the CRISPR RNA (crRNA) of the complex and includes six protein subunits of unknown functions. Using reconstituted Pyrococcus furiosus Cmr complexes, we found that each of the six Cmr proteins plays a critical role in either crRNA interaction or target RNA capture. Cmr2, Cmr3, Cmr4, and Cmr5 are all required for formation of a crRNA-containing complex detected by native gel electrophoresis, and the conserved 5' repeat sequence tag and 5'-OH group of the crRNA are essential for the interaction. Interestingly, capture of the complementary target RNA additionally requires both Cmr1 and Cmr6. In detailed functional studies, we determined that P. furiosus Cmr complexes cleave target RNAs at 6-nucleotide (nt) intervals in the region of complementarity, beginning 5 nt downstream from the crRNA tag and continuing to within ∼14 nt of the 3' end of the crRNA. Our findings indicate that Cmr3 recognizes the signature crRNA tag sequence (and depends on protein-protein interactions with Cmr2, Cmr4, and Cmr5), each Cmr4 subunit mediates a target RNA cleavage, and Cmr1 and Cmr6 mediate an essential interaction between the 3' region of the crRNA and the target RNA.
Keywords: CRISPR; Cas; Cmr; Pyrococcus furiosus; RNAi; crRNA.
© 2014 Hale et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Figure 1.
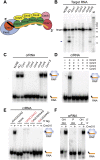
Requirements for target RNA cleavage and crRNA binding by the Cmr complex. (A) Model of the organization of the Cmr protein subunits relative to the crRNA within the Cmr complex (Spilman et al. 2013). (B) Cmr protein requirements for target RNA cleavage. The indicated recombinant Cmr proteins were preincubated with the 45-nt crRNA followed by addition of 5′ end-labeled complementary target RNA. The resulting products were separated by denaturing gel electrophoresis. Radiolabeled Decade marker RNAs (M) were used for size determination. The major target RNA cleavage product is indicated (arrow). (C) Cmr protein requirements for crRNA binding. The indicated recombinant Cmr proteins were incubated with 3′ end-labeled 45-nt crRNA. RNA and complexes were resolved by native gel electrophoresis. (D) Cmr protein requirements for formation of the Cmr2–5 crRNP. The indicated Cmr proteins (Cmr2–5) were incubated with 3′ end-labeled 45-nt crRNA, and complex formation was analyzed by native gel electrophoresis. (E) 5′ tag sequence requirements for formation of the Cmr2–5 crRNP. Recombinant Cmr2–5 proteins were incubated with crRNAs that contain either a wild-type (AUUGAAAG) 5′ repeat tag, no tag, or a mutated tag sequence (complete substitution, addition of 5′ G nucleotide, or substitution of first 2 nt of 8 nt; indicated in red), and complex formation was analyzed by native gel electrophoresis. (F) End group requirements for formation of the Cmr2–5 crRNP. Recombinant Cmr2–5 proteins were incubated with crRNAs containing the indicated 5′ and 3′ chemical end groups, and complex formation was analyzed by native gel electrophoresis. Diagrams indicate the positions of unbound crRNA and crRNPs (in the absence and presence of proteins, respectively) in C_–_F.
Figure 2.
Cmr protein–crRNA contacts in the Cmr2–5 complex. (A) UV cross-linking substrates used to analyze the locations of Cmr protein–crRNA contacts in the Cmr2–5 subcomplex. Labeled crRNAs included in preparations used for UV cross-linking assays (1–7) are diagrammed with the tag and guide regions delineated. Yellow segments indicate regions containing radiolabeled nucleotides. (B) Electrophoretic positions of Cmr proteins (left panel) and cross-linked radiolabeled nucleotides (right panel) following UV cross-linking. crRNA substrates diagrammed in A (1–7) were incubated with Cmr proteins 2–5 and exposed to UV light. The complexes were denatured and subjected to digestion by either RNase T1 or A. (Left panel) The samples were separated on SDS-PAGE gels, followed by Coomassie staining. The positions of the Cmr proteins and the RNases are indicated at the left. The crRNA substrates are indicated at the bottom of the lane. (Right panel) The Coomassie-stained gel (shown in the left panel) was subjected to phosphorimaging. The positions of the Cmr proteins with observable cross-linking to crRNAs are indicated. The asterisk indicates a nonspecific protein observed in all reactions. (C) Summary of observed Cmr protein–crRNA cross-links. The regions of the crRNA where the indicated Cmr subunits were observed to cross-link are illustrated.
Figure 3.
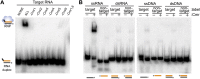
Requirements for recognition of the 5′ repeat-derived crRNA tag sequence. In all panels, the indicated recombinant Cmr proteins were incubated with an 8-nt tag RNA (modified where indicated and generally 3′ end-labeled unless noted). RNA and complexes were resolved by native gel electrophoresis. Diagrams indicate the positions of unbound crRNA and crRNPs (in the absence and presence of proteins, respectively). (A) Individual Cmr proteins required for 5′ tag RNA binding by the full complex. (B) Individual Cmr proteins required for 5′ tag RNA binding by the Cmr2–5 subcomplex. (C) Sequence required for 5′ tag RNA binding by the Cmr2–5 subcomplex. Wild-type (AUUGAAAG) and mutated (UAACUUUC) tag RNAs were analyzed as indicated. (D) End groups required for 5′ tag RNA binding by the Cmr2–5 subcomplex. See the Materials and Methods for details about preparation of RNAs with the indicated 5′ and 3′ chemical end groups.
Figure 4.
Requirements for target binding by the Cmr complex. (A) Cmr protein requirements for target RNA binding. The indicated recombinant Cmr proteins and unlabeled 45-nt crRNA were combined and incubated with 5′ end-labeled complementary target RNA. Complex formation was analyzed by native gel electrophoresis. (B) Target binding specificity. Recombinant Cmr1–6 and unlabeled 45-nt crRNA were combined and incubated with the indicated radiolabeled RNA or DNA substrates. Substrates are illustrated below each lane; asterisk indicates label location and substrate strand. “Target” substrates (gray) are complementary to the crRNA. “Nontarget” substrates (orange) are not complementary (equivalent to guide sequence of crRNA). Diagrams at the left of the panel indicate the electrophoretic positions of unbound labeled substrate (in the presence of crRNA) and crRNPs (in the presence of proteins).
Figure 5.
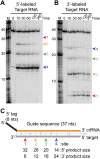
Analysis of cleavage of 5′- and 3′-labeled target RNAs. (A,B) Cleavage of 5′- and 3′-labeled target RNAs by the Cmr complex. Recombinant Cmr1–6 proteins and unlabeled 45-nt crRNA were combined and incubated with 5′-labeled (A) or 3′-labeled (B) complementary target RNAs. Samples were incubated for the indicated amount of time at 70°C or for 360 min on ice (ice), and the labeled target RNAs were separated by high-resolution denaturing polyacrylamide gel electrophoresis. Colored arrows indicate the products of cleavage at the four observed cleavage sites. Radiolabeled Decade marker RNAs (M) were used for size determination. The dotted line indicates noncontiguous lanes. (C) Summary of the mapped target RNA cleavage sites. The target RNA (gray) is fully complementary to the guide region (orange) of the crRNA used in the assay. Colored arrows indicate the positions of the observed cleavages, corresponding to the products observed in A and B. The sizes of the products observed in reactions with 5′- and 3′-labeled substrates are indicated below, and distances between observed cleavage sites are shown in gray. The 3′ end-labeled target RNA is generated by ligation of a radiolabeled pCp moiety, which adds a single nucleotide to the 3′ end of the target RNA (indicated with a box).
Figure 6.
Analysis of cleavages directed by crRNAs of variable guide sequence length. (A,B) Cleavage directed by Cmr complexes containing crRNAs with a series of shorter guide sequences. Recombinant Cmr1–6 and unlabeled crRNAs of the indicated lengths were combined and incubated with 5′ end-labeled (A) or 3′ end-labeled (B) complementary target RNA. (R) Target RNA alone; (−) no crRNA was added. Cleavage products were separated by high-resolution denaturing polyacrylamide gel electrophoresis. Colored arrows indicate the products of cleavage at the four observed cleavage sites (labeled as in Fig. 5). The doublet observed for the smallest product of the 3′-labeled target RNA (cleavage site 1) is likely due to partial loss of the 3′ phosphate added during the 3′ pCp labeling reaction. (B) Cleavages by Cmr complexes containing a crRNA with an extended guide sequence. Recombinant Cmr1–6 and unlabeled 67-nt crRNAs were combined and incubated with 5′ end-labeled (C) or 3′ end-labeled (D) complementary target RNA. The reactions were incubated for the indicated amount of time at 70°C or for 360 minutes on ice (ice). (Target RNA alone [R] was incubated for 360 minutes at 70°C.) Colored arrows indicate the products of cleavage at the eight observed cleavage sites. In A_–_D, radiolabeled Decade marker RNAs (M) were used for size determination. (E) Summary of the mapped target RNA cleavage sites. Sites of cleavage guided by crRNAs of various lengths are indicated by colored arrows and correspond to the products observed in A_–_D. The length of the crRNA guide region is indicated in parentheses, and the distances from the ends of the guide region to the nearest site of cleavage are indicated.
Figure 7.
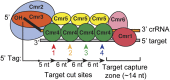
Functional organization of the Cmr complex. The organization of the Cmr proteins along the length of the crRNA in the P. furiosus Cmr complex is shown (Spilman et al. 2013). The findings presented here provide significant functional insights on this complex. Four of the Cmr proteins—Cmr2–5 (blue, orange, green, and yellow)—are required to specifically bind RNAs with the signature CRISPR tag sequence (Fig. 1); Cmr3 interacts with the tag directly (Fig. 2; Spilman et al. 2013). Target RNA capture additionally requires Cmr1 and Cmr6 (red and pink) (Fig. 4). Cmr complexes cleave target RNAs at multiple sites located at 6-nt intervals in the region of complementarity, beginning 5 nt downstream from the crRNA tag (Figs. 5, 6). Cleavages can occur up through a region of ∼14 nt at the end of the target RNA–crRNA duplex. This target capture zone is critical for target RNA cleavage (Hale et al. 2009) and is the binding site of Cmr1 and Cmr6 (Spilman et al. 2013), which are critical for target RNA capture (Fig. 4). Our data indicate that Cmr4 plays a critical role in specifying the target RNA cleavages and may be the catalytic component of the Cmr complex.
Similar articles
- Structure of an RNA silencing complex of the CRISPR-Cas immune system.
Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Spilman M, et al. Mol Cell. 2013 Oct 10;52(1):146-52. doi: 10.1016/j.molcel.2013.09.008. Mol Cell. 2013. PMID: 24119404 Free PMC article. - Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog.
Osawa T, Inanaga H, Sato C, Numata T. Osawa T, et al. Mol Cell. 2015 May 7;58(3):418-30. doi: 10.1016/j.molcel.2015.03.018. Epub 2015 Apr 23. Mol Cell. 2015. PMID: 25921071 - Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex.
Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, Li H, Stagg SM. Ramia NF, et al. Cell Rep. 2014 Dec 11;9(5):1610-1617. doi: 10.1016/j.celrep.2014.11.007. Epub 2014 Dec 4. Cell Rep. 2014. PMID: 25482566 Free PMC article. - The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus.
Terns RM, Terns MP. Terns RM, et al. Biochem Soc Trans. 2013 Dec;41(6):1416-21. doi: 10.1042/BST20130056. Biochem Soc Trans. 2013. PMID: 24256230 Free PMC article. Review. - Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus.
Zhang J, White MF. Zhang J, et al. Biochem Soc Trans. 2013 Dec;41(6):1422-6. doi: 10.1042/BST20130031. Biochem Soc Trans. 2013. PMID: 24256231 Review.
Cited by
- Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex.
Zhu X, Ye K. Zhu X, et al. Nucleic Acids Res. 2015 Jan;43(2):1257-67. doi: 10.1093/nar/gku1355. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25541196 Free PMC article. - Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence.
Grüschow S, Athukoralage JS, Graham S, Hoogeboom T, White MF. Grüschow S, et al. Nucleic Acids Res. 2019 Sep 26;47(17):9259-9270. doi: 10.1093/nar/gkz676. Nucleic Acids Res. 2019. PMID: 31392987 Free PMC article. - SCOPE enables type III CRISPR-Cas diagnostics using flexible targeting and stringent CARF ribonuclease activation.
Steens JA, Zhu Y, Taylor DW, Bravo JPK, Prinsen SHP, Schoen CD, Keijser BJF, Ossendrijver M, Hofstra LM, Brouns SJJ, Shinkai A, van der Oost J, Staals RHJ. Steens JA, et al. Nat Commun. 2021 Aug 19;12(1):5033. doi: 10.1038/s41467-021-25337-5. Nat Commun. 2021. PMID: 34413302 Free PMC article. - Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers.
Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. Niewoehner O, et al. Nature. 2017 Aug 31;548(7669):543-548. doi: 10.1038/nature23467. Epub 2017 Jul 19. Nature. 2017. PMID: 28722012 - Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex.
Blosser TR, Loeff L, Westra ER, Vlot M, Künne T, Sobota M, Dekker C, Brouns SJJ, Joo C. Blosser TR, et al. Mol Cell. 2015 Apr 2;58(1):60-70. doi: 10.1016/j.molcel.2015.01.028. Epub 2015 Mar 5. Mol Cell. 2015. PMID: 25752578 Free PMC article.
References
- Bailey S. 2013. The Cmr complex: an RNA-guided endoribonuclease. Biochem Soc Trans 41: 1464–1467 - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM066958/GM/NIGMS NIH HHS/United States
- R01 GM054682/GM/NIGMS NIH HHS/United States
- R01 GM099604/GM/NIGMS NIH HHS/United States
- R01GM54682/GM/NIGMS NIH HHS/United States
- R01GM99604/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials