ANGSD: Analysis of Next Generation Sequencing Data - PubMed (original) (raw)
ANGSD: Analysis of Next Generation Sequencing Data
Thorfinn Sand Korneliussen et al. BMC Bioinformatics. 2014.
Abstract
Background: High-throughput DNA sequencing technologies are generating vast amounts of data. Fast, flexible and memory efficient implementations are needed in order to facilitate analyses of thousands of samples simultaneously.
Results: We present a multithreaded program suite called ANGSD. This program can calculate various summary statistics, and perform association mapping and population genetic analyses utilizing the full information in next generation sequencing data by working directly on the raw sequencing data or by using genotype likelihoods.
Conclusions: The open source c/c++ program ANGSD is available at http://www.popgen.dk/angsd . The program is tested and validated on GNU/Linux systems. The program facilitates multiple input formats including BAM and imputed beagle genotype probability files. The program allow the user to choose between combinations of existing methods and can perform analysis that is not implemented elsewhere.
Figures
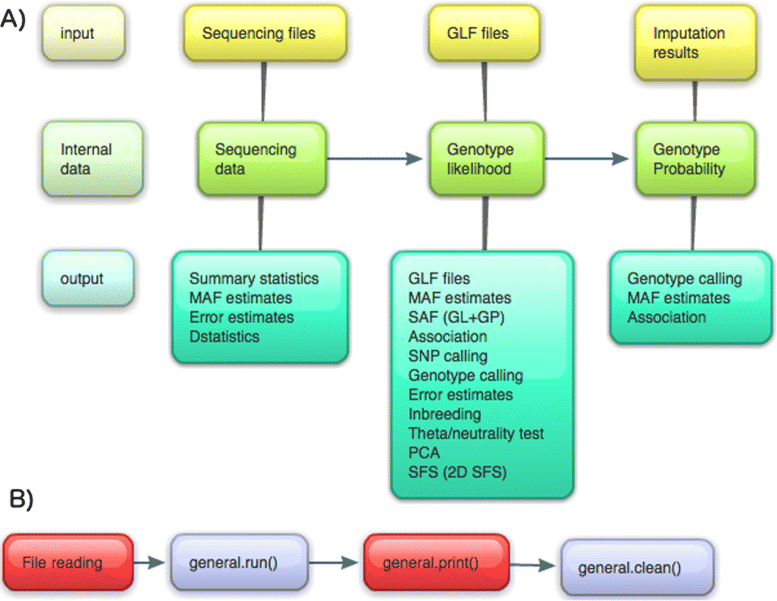
Figure 1
Data formats and call graph. A) Dependency of different data formats and analyses that can be performed in ANGSD. B) Simplified call graph. Red nodes indicate areas that are not threaded. With the exception of file readers, all analyses, printing and cleaning is done by objects derived from the abstract base class called general.
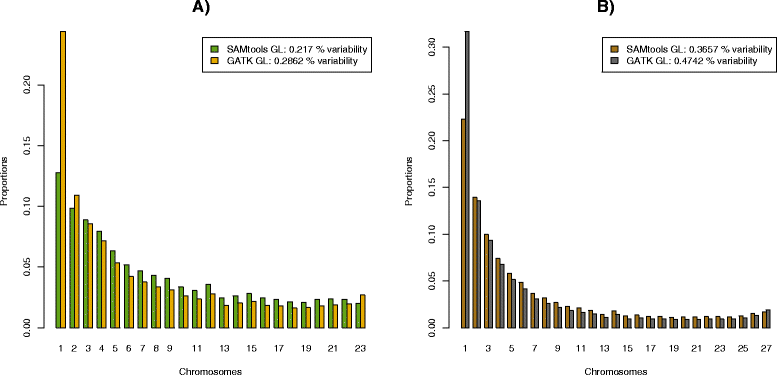
Figure 2
1D SFS for different GL models. SFS estimation based on a 170 megabase region from chromosome 1 using 12 CEU samples A) and 14 YRI samples **B)**” from the 1000 genomes project. The analysis was performed for both the GATK GL model (green, light brown) and SAMtools GL (yellow,dark brown). Notice the difference in estimated variability (proportion of variable sites) for the two GL models, with GATK GL based analyses inferring more variable sites and an associated larger proportion of low-frequency alleles. The two categories of invariable sites have been removed and the distributions have been normalized so that the frequencies of all categories sum to one for each method.
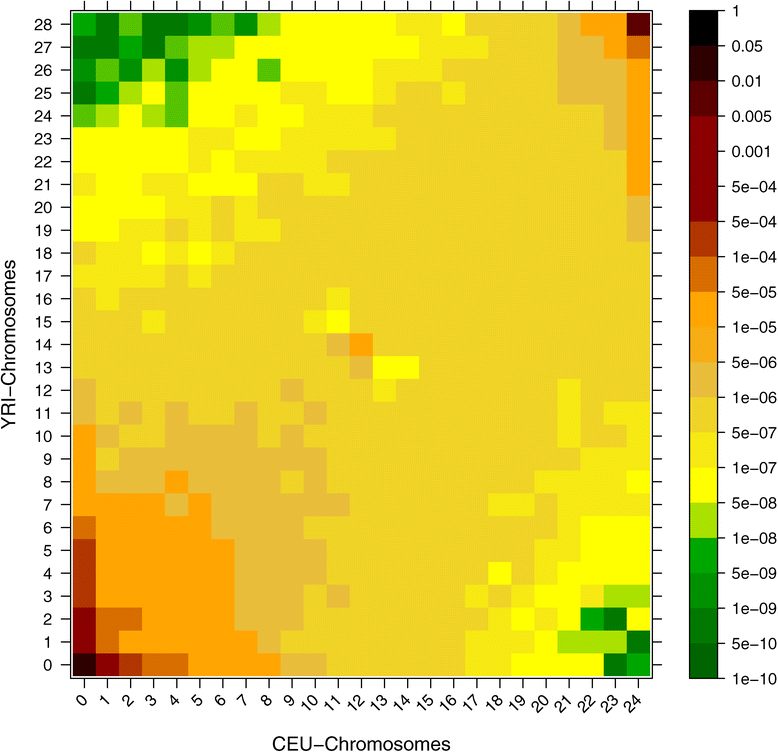
Figure 3
Joint SFS (2D-SFS). Two dimensional SFS estimation based on a 170 megabase region from chromosome 1 using 12 CEU samples and 14 YRI samples from the 1000 genomes project.
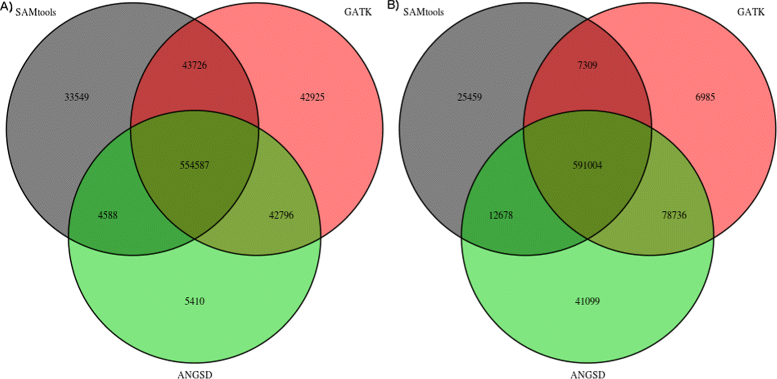
Figure 4
Overlap between inferred SNPs with a critical p-value threshold of 10 −6 and not using BAQ. Venn diagram of the overlap between the SNP discovery for ANGSD, GATK and SAMtools for 33 CEU samples for chromosome 1. We used default parameters with GATK for SAMtools we discarded reads with a mapping quality below 10. For ANGSD we choose an p-value threshold of 10−6 and didn’t enable BAQ. In A, we used the SAMtools genotype likelihood model in ANGSD, in B we used the GATK model in ANGSD.
Figure 5
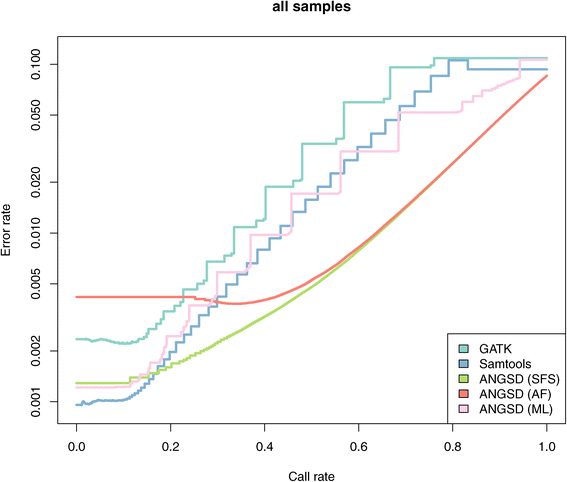
Error rate vs call rate for called genotypes. Error rate and call rates for genotype calls based on different methods. The error rate is defined as the discordance rate between HapMap genotype calls compared to the same individuals sequenced in the 1000 genomes. Genotype where called for all sites for all individuals for all methods. Each genotype call has a score which was used to determine the call rate. Due to the discrete nature of some of the genotype scores we obtain a jagged curve.
Similar articles
- angsd-wrapper: utilities for analysing next-generation sequencing data.
Durvasula A, Hoffman PJ, Kent TV, Liu C, Kono TJ, Morrell PL, Ross-Ibarra J. Durvasula A, et al. Mol Ecol Resour. 2016 Nov;16(6):1449-1454. doi: 10.1111/1755-0998.12578. Epub 2016 Aug 29. Mol Ecol Resour. 2016. PMID: 27480660 - Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding.
Hanghøj K, Moltke I, Andersen PA, Manica A, Korneliussen TS. Hanghøj K, et al. Gigascience. 2019 May 1;8(5):giz034. doi: 10.1093/gigascience/giz034. Gigascience. 2019. PMID: 31042285 Free PMC article. - fastNGSadmix: admixture proportions and principal component analysis of a single NGS sample.
Jørsboe E, Hanghøj K, Albrechtsen A. Jørsboe E, et al. Bioinformatics. 2017 Oct 1;33(19):3148-3150. doi: 10.1093/bioinformatics/btx474. Bioinformatics. 2017. PMID: 28957500 - Estimating individual admixture proportions from next generation sequencing data.
Skotte L, Korneliussen TS, Albrechtsen A. Skotte L, et al. Genetics. 2013 Nov;195(3):693-702. doi: 10.1534/genetics.113.154138. Epub 2013 Sep 11. Genetics. 2013. PMID: 24026093 Free PMC article. - NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data.
Korneliussen TS, Moltke I. Korneliussen TS, et al. Bioinformatics. 2015 Dec 15;31(24):4009-11. doi: 10.1093/bioinformatics/btv509. Epub 2015 Aug 30. Bioinformatics. 2015. PMID: 26323718 Free PMC article.
Cited by
- The genomic portrait of the Picene culture provides new insights into the Italic Iron Age and the legacy of the Roman Empire in Central Italy.
Ravasini F, Kabral H, Solnik A, de Gennaro L, Montinaro F, Hui R, Delpino C, Finocchi S, Giroldini P, Mei O, Beck De Lotto MA, Cilli E, Hajiesmaeil M, Pistacchia L, Risi F, Giacometti C, Scheib CL, Tambets K, Metspalu M, Cruciani F, D'Atanasio E, Trombetta B. Ravasini F, et al. Genome Biol. 2024 Nov 21;25(1):292. doi: 10.1186/s13059-024-03430-4. Genome Biol. 2024. PMID: 39567978 Free PMC article. - Ancient genomes from the Tang Dynasty capital reveal the genetic legacy of trans-Eurasian communication at the eastern end of Silk Road.
Lv M, Ma H, Wang R, Li H, Zhang X, Zhang W, Zeng Y, Qin Z, Zhai H, Lou Y, Lin Y, Tao L, He H, Yang X, Zhu K, Zhou Y, Wang CC. Lv M, et al. BMC Biol. 2024 Nov 20;22(1):267. doi: 10.1186/s12915-024-02068-9. BMC Biol. 2024. PMID: 39567925 Free PMC article. - Sequential introgression of a carotenoid processing gene underlies sexual ornament diversity in a genus of manakins.
Lim HC, Bennett KFP, Justyn NM, Powers MJ, Long KM, Kingston SE, Lindsay WR, Pease JB, Fuxjager MJ, Bolton PE, Balakrishnan CN, Day LB, Parsons TJ, Brawn JD, Hill GE, Braun MJ. Lim HC, et al. Sci Adv. 2024 Nov 22;10(47):eadn8339. doi: 10.1126/sciadv.adn8339. Epub 2024 Nov 20. Sci Adv. 2024. PMID: 39565864 Free PMC article. - Prioritizing Conservation Areas for the Hyacinth Macaw (Anodorhynchus hyacinthinus) in Brazil From Low-Coverage Genomic Data.
Vilaça ST, Dalapicolla J, Soares R, Guedes NMR, Miyaki CY, Aleixo A. Vilaça ST, et al. Evol Appl. 2024 Nov 18;17(11):e70039. doi: 10.1111/eva.70039. eCollection 2024 Nov. Evol Appl. 2024. PMID: 39564451 Free PMC article. - Historical and ongoing hybridisation in Southern South American grassland species.
Giudicelli GC, Pezzi PH, Guzmán-Rodriguez S, Turchetto C, Bombarely A, Freitas LB. Giudicelli GC, et al. Sci Rep. 2024 Nov 14;14(1):27989. doi: 10.1038/s41598-024-79584-9. Sci Rep. 2024. PMID: 39543384 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous