Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma - PubMed (original) (raw)
Oscar Krijgsman 1, Jennifer Tsoi 2, Lidia Robert 3, Willy Hugo 3, Chunying Song 3, Xiangju Kong 3, Patricia A Possik 1, Paulien D M Cornelissen-Steijger 1, Marnix H Geukes Foppen 4, Kristel Kemper 1, Colin R Goding 5, Ultan McDermott 6, Christian Blank 4, John Haanen 4, Thomas G Graeber 7, Antoni Ribas 8, Roger S Lo 8, Daniel S Peeper 1
Affiliations
- PMID: 25502142
- PMCID: PMC4428333
- DOI: 10.1038/ncomms6712
Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma
Judith Müller et al. Nat Commun. 2014.
Abstract
Increased expression of the Microphthalmia-associated transcription factor (MITF) contributes to melanoma progression and resistance to BRAF pathway inhibition. Here we show that the lack of MITF is associated with more severe resistance to a range of inhibitors, while its presence is required for robust drug responses. Both in primary and acquired resistance, MITF levels inversely correlate with the expression of several activated receptor tyrosine kinases, most frequently AXL. The MITF-low/AXL-high/drug-resistance phenotype is common among mutant BRAF and NRAS melanoma cell lines. The dichotomous behaviour of MITF in drug response is corroborated in vemurafenib-resistant biopsies, including MITF-high and -low clones in a relapsed patient. Furthermore, drug cocktails containing AXL inhibitor enhance melanoma cell elimination by BRAF or ERK inhibition. Our results demonstrate that a low MITF/AXL ratio predicts early resistance to multiple targeted drugs, and warrant clinical validation of AXL inhibitors to combat resistance of BRAF and NRAS mutant MITF-low melanomas.
Figures
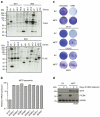
Figure 1. Overexpressed MITF protects cells against ERK inhibition
(a) An insertional mutagenesis screen was performed using the VBIM system. A BRAF mutant melanoma cell line of low passage (04.07) was infected with three VBIM vectors (SD1–3; to cover the three reading frames) and treated with 1 μM of the ERK inhibitor SCH772984. Drug-resistant clones were established and analysed for FLAG-tagged proteins resulting from the integration of the viral cassettes. (b) Integration of the VBIM cassettes resulted in MITF overexpression in six independent ERKi-resistant clones, as shown by qRT–PCR, compared with parental 04.07 cells. Note that only clones derived from separate plates were considered as independent clones. Error bars denote s.d. for technical replicates. (c) Lentiviral overexpression of MITF resulted in increased resistance to long-term ERK inhibition in three independent BRAF mutant melanoma cell lines as shown by crystal violet staining. (d) Immunoblot of 04.07 cells confirmed MITF overexpression and inactivation of ERK after inhibitor treatment. HSP90 served as a loading control.
Figure 2. MITF loss occurs in acquired resistance in vitro and in vivo
(a) Treatment-naïve BRAF mutant melanoma cells were under constant treatment with PLX4720 until resistance was achieved. Crystal violet staining confirmed resistance to 3 μM PLX4720 in resistant cells in comparison with their sensitive counterparts. (b) Immunoblot of sensitive (S) and resistant (R) melanoma cells showed expression of MITF and MITF-regulated proteins. HSP90 served as a loading control. (c) MITF mRNA expression was measured by qRT–PCR in treatment-naïve and resistant melanoma cell lines normalized to RPL13 mRNA expression. Error bars denote s.d. for technical replicates. (d) Human melanoma biopsies (obtained from patients before vemurafenib treatment (pre) and after relapse had occurred (post)) were stained for MITF. Tumour 2 was from a patient with distinct relapsed tumour clones. Scale bars, 200 μm. (e) _MITF_-M mRNA was measured in human biopsies before vemurafenib treatment (pre) and after relapse had occurred (post) from two different patients. Values were normalized to GAPDH mRNA expression. P values, by paired Student’s _t_-test across triplicates, were only shown for samples for which MITF mRNA expression dropped. Error bars denote s.d. for technical triplicates.
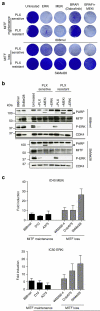
Figure 3. MITF loss causes cross-resistance to MAPK pathway inhibition
(a) Two PLX4720-resistant cell lines (888mel and SkMel28) and their treatment-naïve counterparts were treated with either the ERKi SCH772984 (0.5 μM), the MEKi trametinib (0.3 μM), BRAF inhibitor dabrafenib (3 μM) or a combination as indicated and stained with crystal violet after 6 days of treatment. (b) Immunoblotting for the indicated proteins confirmed the downregulation of the MAPK pathway (as judged by P-ERK) and showed cleaved PARP (arrow) as a measure of apoptosis of the cell lines used in a. CDK4 served as a loading control. (c) Six PLX4720-resistant cell lines and their treatment-naïve counterparts were separated in groups that either lose or maintain (or increase) MITF expression on acquiring resistance. These cells were exposed to the MEKi trametinib or the ERKi SCH772984 in dose–response curves. IC40 (for MEKi) or IC50 (for ERKi) were calculated from the curves. Fold increase in IC40 or IC50 was determined comparing resistant to sensitive cells. Mean was calculated from three independent experiments, error bars indicate s.d.
Figure 4. Loss of MITF is accompanied by increased invasiveness
(a) PLX4720-resistant melanoma cells and their treatment-naïve counterparts were seeded on a matrigel-coated invasion chamber for 12 h. Cells that invaded through the matrix were stained with crystal violet. Scale bars, 500 μm. (b) Quantification of a from at least five images. (c) PLX4720-resistant cells (R) and their treatment-naïve (S) counterparts were blotted for EMT markers. α-Tubulin served as a loading control. (d) ZEB1 and ZEB2 mRNA were measured in acquired PLX4720-resistant cells and their treatment-naïve counterparts. The results are based on three independent experiments and normalized to RPL13. Data are shown as mean + s.d. (e) mRNA expression of Dia1 was measured in acquired PLX4720-resistant cells and their treatment-naïve counterparts. The results are based on three independent experiments and normalized to RPL13. Data are shown as mean + s.d. (f) Knockdown of Fra-1 in two PLX4720-resistant cell lines was documented on immunoblot. HSP90 served as a loading control. (g) Two PLX4720-resistant melanoma cells transduced with Fra-1 knockdown and control cells were seeded on a matrigel-coated invasion chamber for 12 h. Cells that invaded through the matrix were stained with crystal violet. Inserts show proliferation in 12 h on polystyrene culture dishes. Scale bars, 200 μm. (h) Quantification of g of at least five images.
Figure 5. Absence of MITF marks intrinsically insensitive cells
(a) Treatment-naïve BRAF mutant melanoma cells were grouped based on their MITF expression by immunoblotting, also for additional proteins as indicated. HSP90 served as a loading control. (b) BRAF mutant melanoma cells were plated at low density and treated with 5 μM PLX4720 for 6 days or left untreated and stained with crystal violet. (c) For a subset of the cells lines from b, MAPK pathway inhibition and apoptosis indicated by cleaved PARP (arrow) after 3 days of treatment with 5 μM PLX4720 was confirmed by immunoblotting. HSP90 served as a loading control. (d) Sensitivity of MITFendo_hi and MITFendo_lo treatment-naïve melanoma cell lines to the BRAFi PLX4720, to the ERKi SCH772984 and to the MEKi trametinib was determined in dose–response curves. Mean was calculated from three independent experiments, error bars indicate s.d. (e) MITFendo_hi and MITFendo_lo BRAFV600E mutant melanoma cell lines were plated at low density and treated with either BRAFi (2 μM), MEKi trametinib (0.1 μM) or a combination. After 6 days, cells were stained with crystal violet. (f) An independent set of treatment-naïve BRAFV600E mutant melanoma cell lines was grouped based on MITF expression, and resistance to the BRAFi (vemurafenib) and the ERKi SCH772984 was determined by dose–response curves. Cell lines with MITF amplification are marked with an asterix.
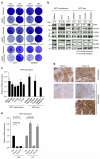
Figure 6. AXL and other RTKs are inversely correlated with MITF
(a) RNA sequencing was performed in three MITFendo_hi and three MITFendo_lo BRAFV600E mutant melanoma cell lines. The genes in the heatmap were selected based on the MITF expression, expression of MITF-target genes and expression of the RTKs AXL, EGFR and PDGFRβ. (b) A phospho-RTK array was performed comparing one MITFendo_hi and one MITFendo_lo melanoma cell line untreated or treated with 5 μM PLX4720 for 2 days. (c) MITFendo_hi and MITFendo_lo cell lines were immunoblotted for the RTKs PDGFRβ, AXL and EGFR. HSP90 served as a loading control. (d) Protein extracts from treatment-naïve BRAF mutant PDX tumours were immunoblotted for the indicated antibodies. (e) Analysis of 356 TCGA melanoma transcriptomes confirming anticorrelation between MITF and RTKs including AXL. P values were computed by a one-sided Wilcoxon ranksum test on the expression levels of AXL, EGFR and PDGFRB between the top quartile (MITF high, n = 89) and bottom quartile (MITF low, n = 89) of MITF expression across the samples.
Figure 7. RTK inhibition increases effect of BRAF inhibition
(a) MITFendo_lo cells were exposed to a combination of RTK inhibitors (AXLi R428 (0.3 μM), EGFRi Gefitinib (2 μM) and PDGFRβi Imatinib (1 μM)) and BRAFi PLX4720 (5 μM). After 9 days of combined treatment the remaining cells were stained with crystal violet. (b) Quantification of a, comprising three independent experiments. Values were normalized to untreated control (100%). Error bars represent s.d.; paired _t_-test was performed for statistical analysis, *P<0.05, **P<0.01, ***P<0.001. (c) PLX4720-resistant melanoma cells and their treatment-naïve counterparts were immunoblotted for AXL, EGFR and PDGFRβ. CDK4 served as a loading control. (d) PLX4720-resistant SkMel28 and their naïve counterparts were blotted on a phospho-RTK array. (e) Matched human melanoma biopsies obtained from a patient before vemurafenib treatment (pre) and after relapse had occurred (post) were stained for MITF and AXL. Scale bars, 100 μm. (f) AXL-expressing PLX4720-resistant melanoma cells were exposed to 0.3 μM of the AXL inhibitor R428 for 9 days and the remaining cells stained with crystal violet.
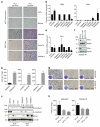
Figure 8. MITF is a predictive biomarker also in NRAS mutant melanomas
(a) RNA sequencing was performed on four MITFendo_hi and four MITFendo_lo NRAS mutant melanoma cell lines. The genes in the heatmap were selected based on the MITF expression, expression of _MITF_-target genes and expression of the mRNAs coding for the RTKs AXL, EGFR and PDGFRβ. (b) Treatment-naïve NRAS mutant melanoma cells were immunoblotted for MITF, RTKs and other proteins as indicated. HSP90 served as a loading control. WM1366-A/B refers to different batches of the same cell line. (c) A phospho-RTK array was performed comparing one NRAS mutant MITFendo_hi and one MITFendo_lo melanoma cell line. (d,e) MITFendo_hi and MITFendo_lo NRAS mutant melanoma cells were plated at low densities and treated with the MEKi trametinib (2.5 nM) (d) or ERKi SCH772984 (0.25 μM) (e) or left untreated for 6 days and stained by crystal violet. (f) Melanoma cells were plated at low density and treated with the AXLi R428 (0.5 μM), the ERKi SCH772984 (1 μM) or in combination. After 6 days of treatment, plates were stained with crystal violet. (g) Quantification of f, based on three biological replicates. Error bars represent s.d.; paired _t_-test was used for statistical analysis, *P<0.05. (h) A MEKi-resistant NRAS mutant melanoma cell line and its treatment-naïve counterpart were blotted for the indicated proteins. HSP90 served as a loading control.
Comment in
- The UPs and DOWNs of MITF in melanoma resistance.
Kim H, Ronai ZA. Kim H, et al. Pigment Cell Melanoma Res. 2015 Mar;28(2):132-3. doi: 10.1111/pcmr.12338. Epub 2014 Dec 23. Pigment Cell Melanoma Res. 2015. PMID: 25476804 No abstract available.
Similar articles
- Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib.
Acquaviva J, Smith DL, Jimenez JP, Zhang C, Sequeira M, He S, Sang J, Bates RC, Proia DA. Acquaviva J, et al. Mol Cancer Ther. 2014 Feb;13(2):353-63. doi: 10.1158/1535-7163.MCT-13-0481. Epub 2014 Jan 7. Mol Cancer Ther. 2014. PMID: 24398428 - BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells.
Lunavat TR, Cheng L, Einarsdottir BO, Olofsson Bagge R, Veppil Muralidharan S, Sharples RA, Lässer C, Gho YS, Hill AF, Nilsson JA, Lötvall J. Lunavat TR, et al. Proc Natl Acad Sci U S A. 2017 Jul 18;114(29):E5930-E5939. doi: 10.1073/pnas.1705206114. Epub 2017 Jul 6. Proc Natl Acad Sci U S A. 2017. PMID: 28684402 Free PMC article. - Mutational activation of BRAF confers sensitivity to transforming growth factor beta inhibitors in human cancer cells.
Spender LC, Ferguson GJ, Liu S, Cui C, Girotti MR, Sibbet G, Higgs EB, Shuttleworth MK, Hamilton T, Lorigan P, Weller M, Vincent DF, Sansom OJ, Frame M, Dijke PT, Marais R, Inman GJ. Spender LC, et al. Oncotarget. 2016 Dec 13;7(50):81995-82012. doi: 10.18632/oncotarget.13226. Oncotarget. 2016. PMID: 27835901 Free PMC article. - Mechanisms of resistance to RAF inhibition in melanomas harboring a BRAF mutation.
Chapman PB. Chapman PB. Am Soc Clin Oncol Educ Book. 2013. doi: 10.14694/EdBook_AM.2013.33.e80. Am Soc Clin Oncol Educ Book. 2013. PMID: 23714462 Review. - Molecularly targeted therapies for melanoma.
Liu LS, Colegio OR. Liu LS, et al. Int J Dermatol. 2013 May;52(5):523-30. doi: 10.1111/j.1365-4632.2012.05829.x. Int J Dermatol. 2013. PMID: 23590367 Review.
Cited by
- DNA promoter hypermethylation of melanocyte lineage genes determines melanoma phenotype.
Sanna A, Phung B, Mitra S, Lauss M, Choi J, Zhang T, Njauw CN, Cordero E, Harbst K, Rosengren F, Cabrita R, Johansson I, Isaksson K, Ingvar C, Carneiro A, Brown K, Tsao H, Andersson M, Pietras K, Jönsson G. Sanna A, et al. JCI Insight. 2022 Oct 10;7(19):e156577. doi: 10.1172/jci.insight.156577. JCI Insight. 2022. PMID: 36040798 Free PMC article. - Spatial and Temporal Changes in PD-L1 Expression in Cancer: The Role of Genetic Drivers, Tumor Microenvironment and Resistance to Therapy.
Shklovskaya E, Rizos H. Shklovskaya E, et al. Int J Mol Sci. 2020 Sep 27;21(19):7139. doi: 10.3390/ijms21197139. Int J Mol Sci. 2020. PMID: 32992658 Free PMC article. Review. - Antibody-drug conjugates: beyond current approvals and potential future strategies.
Menon S, Parakh S, Scott AM, Gan HK. Menon S, et al. Explor Target Antitumor Ther. 2022;3(2):252-277. doi: 10.37349/etat.2022.00082. Epub 2022 Apr 28. Explor Target Antitumor Ther. 2022. PMID: 36046842 Free PMC article. Review. - Cell Adhesion Molecules in Plasticity and Metastasis.
Smart JA, Oleksak JE, Hartsough EJ. Smart JA, et al. Mol Cancer Res. 2021 Jan;19(1):25-37. doi: 10.1158/1541-7786.MCR-20-0595. Epub 2020 Oct 1. Mol Cancer Res. 2021. PMID: 33004622 Free PMC article. Review. - Raman-guided subcellular pharmaco-metabolomics for metastatic melanoma cells.
Du J, Su Y, Qian C, Yuan D, Miao K, Lee D, Ng AHC, Wijker RS, Ribas A, Levine RD, Heath JR, Wei L. Du J, et al. Nat Commun. 2020 Sep 24;11(1):4830. doi: 10.1038/s41467-020-18376-x. Nat Commun. 2020. PMID: 32973134 Free PMC article.
References
- Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous