STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1 - PubMed (original) (raw)
STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1
Katarzyna Blaszczyk et al. Biochem J. 2015.
Abstract
Evidence is accumulating for the existence of a signal transducer and activator of transcription 2 (STAT2)/interferon regulatory factor 9 (IRF9)-dependent, STAT1-independent interferon alpha (IFNα) signalling pathway. However, no detailed insight exists into the genome-wide transcriptional regulation and the biological implications of STAT2/IRF9-dependent IFNα signalling as compared with interferon-stimulated gene factor 3 (ISGF3). In STAT1-defeicient U3C cells stably overexpressing human STAT2 (hST2-U3C) and STAT1-deficient murine embryonic fibroblast cells stably overexpressing mouse STAT2 (mST2-MS1KO) we observed that the IFNα-induced expression of 2'-5'-oligoadenylate synthase 2 (OAS2) and interferon-induced protein with tetratricopeptide repeats 1 (Ifit1) correlated with the kinetics of STAT2 phosphorylation, and the presence of a STAT2/IRF9 complex requiring STAT2 phosphorylation and the STAT2 transactivation domain. Subsequent microarray analysis of IFNα-treated wild-type (WT) and STAT1 KO cells overexpressing STAT2 extended our observations and identified ∼120 known antiviral ISRE-containing interferon-stimulated genes (ISGs) commonly up-regulated by STAT2/IRF9 and ISGF3. The STAT2/IRF9-directed expression profile of these IFN-stimulated genes (ISGs) was prolonged as compared with the early and transient response mediated by ISGF3. In addition, we identified a group of 'STAT2/IRF9-specific' ISGs, whose response to IFNα was ISGF3-independent. Finally, STAT2/IRF9 was able to trigger an antiviral response upon encephalomyocarditis virus (EMCV) and vesicular stomatitis Indiana virus (VSV). Our results further prove that IFNα-activated STAT2/IRF9 induces a prolonged ISGF3-like transcriptome and generates an antiviral response in the absence of STAT1. Moreover, the existence of 'STAT2/IRF9-specific' target genes predicts a novel role of STAT2 in IFNα signalling.
Figures
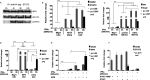
Figure 1. The IFNα response in STAT1 KO cells is abrogated
(A, C) 2fTGH and U3C; (B, D) MEF WT and MS1KO were treated with IFNα for indicated times. For (A) and (B), protein lysates were isolated and analyzed by Western blot analysis. Total STAT2 (tSTAT2), phosphorylated STAT2 (pSTAT2), total STAT1 (tSTAT1), phosphorylated STAT1 (pSTAT1) and IRF9 were analyzed using specific antibodies. Equal loading was verified using anti-tubulin. For (C) and (D), total RNA was extracted and OAS2 and Ifit1 relative fold induction was quantified using qRT-PCR. Statistical significance is presented as compared with the non-treated control (results are means ±S.E.M.). Statistical analysis was conducted using one-way ANOVA with Tukey's post hoc test. *_P_≤0.05, ***_P_≤0.01.
Figure 2. The IFNα response in STAT1 KO cells is recapitulated by increasing STAT2 levels
(A, C) hSTAT2-U3C and Migr1-U3C; (B, D) mST2-MS1KO and Migr1-MS1KO, were treated with IFNα for the indicated times. For (A) and (B), protein lysates were isolated and analyzed by Western blot analysis for expression of tSTAT2, pSTAT2, tSTAT1, pSTAT1 and IRF9. Equal loading was verified using anti-tubulin. In (C, D), total RNA was extracted and OAS2 and Ifit1 relative fold induction was quantified using qRT-PCR. Statistical significance is presented as compared with the non-treated control (results are means±S.E.M.). Statistical analysis was conducted using one-way ANOVA with Tukey's post hoc test. *_P_≤0.05, **_P_≤0.01.
Figure 3. STAT2 and IRF9 complex and mediate an IFNα response in the absence of STAT1
(A) The interaction between STAT2 and IRF9 was analyzed by immunoprecipitation. hSTAT2-U3C were treated with IFNα for the indicated times. Cell lysates were immunoprecipitated with anti-HA antibody followed by Western blotting with IRF9, tSTAT2 and pSTAT2 antibodies. (B) Two different clones of hST2-U3C (hST2-U3Ca and hST2-U3C) varying in hSTAT2 expression level and their control Migr1-U3C; (C) ΔmST2-MS1KO, mST2-MS1KO and their control Migr1-MS1KO; (D) Migr1-U3C, IRF9-U3C and hST2-U3C; (E) hST2-U3C transiently transfected with Migr1-IRF9 (500 ng); (F) U3C cells transiently transfected with STAT2-Y690F or STAT2 plasmid (2.5 μg) were all treated with or without 200 U/ml IFNα for 8 h (B–E) or 24 h (F). Total RNA was extracted and OAS2, Ifit1, STAT2 or IRF9 relative fold inductions were quantified using qRT-PCR. Statistical significance is presented as compared with the non-treated control (results are means ± S.E.M.). Statistical analysis was conducted using one-way ANOVA with Tukey's post hoc test except in (E) where a Student's _t_-test, two-tailed, was used. *_P_≤0.05, **_P_≤0.01.
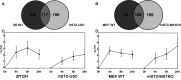
Figure 4. STAT2/IRF9 and ISGF3 regulate expression of a common set of ISGs with different kinetics
(A) 2fTGH and hST2-U3C or (B) MEF WT and mST2-MS1KO were treated with IFNα for 0 h, 4 h, 8 h and 24 h and subjected to microarray analysis. Common up-regulated genes were selected by comparing transcriptomes of individual cell lines. Statistically significant up-regulated genes in human (A) and mouse (B) cell-line data sets were compared by Venn diagram analysis. Average expression profiles of common up-regulated genes between (C) 2fTGH and hST2-U3C and (D) MEF WT and mST2-MS1KO are displayed in centroid view. Expression values are shown as log2 ratio; error bars=S.D.
Figure 5. STAT2/IRF9- and ISGF3-mediated transcriptional responses predict functional overlap
Cluster analysis of common up-regulated genes between (A) 2fTGH and hST2-U3C or (B) MEF WT and mST2-MS1KO. Total RNA from IFNα-treated cell lines was analyzed using Illumina Human HT-12 v4 (A) or MouseRef 8v2 (B) microarrays. For microarray analysis, background subtraction and quantile normalization were used, genes with ratio ≥2 and _P_≤0.05 were considered as up-regulated. log2 ratios from up-regulated genes were clustered using average linkage method.
Figure 6. ChIP-qPCR analysis show enhanced binding of STAT2 to the ISRE of the IFI27, MX1, OAS2, IFIT1, IFIT3 and ISG15 genes in an IFNα-dependent manner in the hST2-U3C cells. Immunoprecipitated DNA was quantified by qPCR and normalized to values obtained after amplification of unprecipitated (input) DNA
ChIP- qPCR confirms enhanced binding of STAT2 to the ISRE in an IFNα-dependent manner in the absence of STAT1.
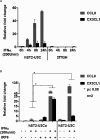
Figure 7. STAT2/IRF9 regulates expression of ISGF3-independent genes
(A) 2fTGH and hST2-U3C were treated with IFNα for indicated times. (B) Two different clones of hST2-U3C (hST2-U3Ca and hST2-U3C) varying in hSTAT2 expression levels were treated with IFNα for 8 h. Subsequently, hST2-U3Ca was transfected with Migr1-IRF9 (500 ng) and treated with IFNα for 8 h. In (A, B), total RNA was extracted. CCL8 and CX3CL1 relative fold induction was quantified using qRT-PCR. All data are presented as means±S.E.M. Statistical significance was assessed using Student's _t_-test, two tailed, *_P_≤0.05.
Figure 8. 2fTGH, U3C, hST2-U3C and Migr1-U3C cell lines, pre-treated for 24 h with 2-fold serial dilutions of IFNα from 250 U/ml, were infected with (A) EMCV or (B) VSV at a MOI of 0.3 for 20 h, or at a MOI of 3 for 20 h (C and D, respectively) followed by visualizing live cells by crystal violet staining
STAT2/IRF9 mediates a similar antiviral response against EMCV and VSV as ISGF3.
Similar articles
- Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections.
Wang W, Yin Y, Xu L, Su J, Huang F, Wang Y, Boor PPC, Chen K, Wang W, Cao W, Zhou X, Liu P, van der Laan LJW, Kwekkeboom J, Peppelenbosch MP, Pan Q. Wang W, et al. Sci Signal. 2017 Apr 25;10(476):eaah4248. doi: 10.1126/scisignal.aah4248. Sci Signal. 2017. PMID: 28442624 - Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9.
Li W, Hofer MJ, Songkhunawej P, Jung SR, Hancock D, Denyer G, Campbell IL. Li W, et al. J Biol Chem. 2017 Apr 7;292(14):5845-5859. doi: 10.1074/jbc.M116.756510. Epub 2017 Feb 17. J Biol Chem. 2017. PMID: 28213522 Free PMC article. - ISGF3 and STAT2/IRF9 Control Basal and IFN-Induced Transcription through Genome-Wide Binding of Phosphorylated and Unphosphorylated Complexes to Common ISRE-Containing ISGs.
Nowicka H, Sekrecka A, Blaszczyk K, Kluzek K, Chang CY, Wesoly J, Lee CK, Bluyssen HAR. Nowicka H, et al. Int J Mol Sci. 2023 Dec 18;24(24):17635. doi: 10.3390/ijms242417635. Int J Mol Sci. 2023. PMID: 38139463 Free PMC article. - The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses.
Blaszczyk K, Nowicka H, Kostyrko K, Antonczyk A, Wesoly J, Bluyssen HA. Blaszczyk K, et al. Cytokine Growth Factor Rev. 2016 Jun;29:71-81. doi: 10.1016/j.cytogfr.2016.02.010. Epub 2016 Mar 18. Cytokine Growth Factor Rev. 2016. PMID: 27053489 Review. - A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.
Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. Michalska A, et al. Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
Cited by
- STAT2 dependent Type I Interferon response promotes dysbiosis and luminal expansion of the enteric pathogen Salmonella Typhimurium.
Wilson RP, Tursi SA, Rapsinski GJ, Medeiros NJ, Le LS, Kotredes KP, Patel S, Liverani E, Sun S, Zhu W, Kilpatrick L, Winter SE, Gamero AM, Tükel Ç. Wilson RP, et al. PLoS Pathog. 2019 Apr 22;15(4):e1007745. doi: 10.1371/journal.ppat.1007745. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 31009517 Free PMC article. - The role of the interferon/JAK-STAT axis in driving islet HLA-I hyperexpression in type 1 diabetes.
Russell MA, Richardson SJ, Morgan NG. Russell MA, et al. Front Endocrinol (Lausanne). 2023 Oct 6;14:1270325. doi: 10.3389/fendo.2023.1270325. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37867531 Free PMC article. Review. - STAT family of transcription factors in breast cancer: Pathogenesis and therapeutic opportunities and challenges.
Wong GL, Manore SG, Doheny DL, Lo HW. Wong GL, et al. Semin Cancer Biol. 2022 Nov;86(Pt 3):84-106. doi: 10.1016/j.semcancer.2022.08.003. Epub 2022 Aug 19. Semin Cancer Biol. 2022. PMID: 35995341 Free PMC article. Review. - Abnormalities of the type I interferon signaling pathway in lupus autoimmunity.
Gallucci S, Meka S, Gamero AM. Gallucci S, et al. Cytokine. 2021 Oct;146:155633. doi: 10.1016/j.cyto.2021.155633. Epub 2021 Jul 30. Cytokine. 2021. PMID: 34340046 Free PMC article. Review. - Targeted Stat2 deletion in conventional dendritic cells impairs CTL responses but does not affect antibody production.
Qiu CC, Kotredes KP, Cremers T, Patel S, Afanassiev A, Slifker M, Gallucci S, Gamero AM. Qiu CC, et al. Oncoimmunology. 2020 Dec 29;10(1):1860477. doi: 10.1080/2162402X.2020.1860477. Oncoimmunology. 2020. PMID: 33457079 Free PMC article.
References
- Bluyssen H.A., Rastmanesh M.M., Tilburgs C., Jie K., Wesseling S., Goumans M.J., Boer P., Joles J.A., Braam B. IFN gamma-dependent SOCS3 expression inhibits IL-6-induced STAT3 phosphorylation and differentially affects IL-6 mediated transcriptional responses in endothelial cells. Am. J. Physiol. Cell Physiol. 2010;299:C354–C362. doi: 10.1152/ajpcell.00513.2009. - DOI - PubMed
- Wesoly J., Szweykowska-Kulinska Z., Bluyssen H.A. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim. Pol. 2007;54:27–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous