A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication - PubMed (original) (raw)
A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication
Peng Wang et al. Nat Med. 2015 Apr.
Abstract
Types 1 and 2 diabetes affect some 380 million people worldwide. Both ultimately result from a deficiency of functional pancreatic insulin-producing beta cells. Beta cells proliferate in humans during a brief temporal window beginning around the time of birth, with a peak percentage (∼2%) engaged in the cell cycle in the first year of life. In embryonic life and after early childhood, beta cell replication is barely detectable. Whereas beta cell expansion seems an obvious therapeutic approach to beta cell deficiency, adult human beta cells have proven recalcitrant to such efforts. Hence, there remains an urgent need for antidiabetic therapeutic agents that can induce regeneration and expansion of adult human beta cells in vivo or ex vivo. Here, using a high-throughput small-molecule screen (HTS), we find that analogs of the small molecule harmine function as a new class of human beta cell mitogenic compounds. We also define dual-specificity tyrosine-regulated kinase-1a (DYRK1A) as the likely target of harmine and the nuclear factors of activated T cells (NFAT) family of transcription factors as likely mediators of human beta cell proliferation and differentiation. Using three different mouse and human islet in vivo-based models, we show that harmine is able to induce beta cell proliferation, increase islet mass and improve glycemic control. These observations suggest that harmine analogs may have unique therapeutic promise for human diabetes therapy. Enhancing the potency and beta cell specificity of these compounds are important future challenges.
Figures
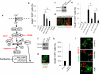
Figure 1. High-throughput screening reveals harmine family members as agonists of beta cell proliferation
(a) Schematic outline of the screen in HepG2 cells used to identify compounds that promote beta cell replication. See text and Online Methods for details. (b) Results of the primary screen showing the 4500 initial hits (black) and the 86 compounds with a median absolute deviation (MAD) score >3 (green). (c) Examples of tertiary screening (rat beta cell BrdU incorporation) of the 86 compounds. Compound 1 is harmine. “D” is DMSO and “C” indicates rat islets treated with no vehicle. The BrdU screen was performed four times; where no error bars are seen, they are within the bar. (d) Examples of BrdU and Ki-67 labeling human beta cells treated with harmine. Note BrdU and Ki-67 nuclear “doublets” in human beta cells. (e) An enlarged view of harmine-treated human beta cells with Ki-67 nuclear “doublets” in adjacent cells. (f) Quantification of BrdU incorporation into rat (left) and human (right) beta cells. “C” indicates control (DMSO, vehicle) and “H” harmine. A minimum of 1000 beta cells was counted from multiple donors (4 rat, 6 human) for each bar. (g) Quantification of Ki67 labeling in rat and human beta cells. “C” indicates control (vehicle, DMSO) and “H” harmine. A minimum of 1000 beta cells was counted from multiple donor pairs (4 rat, 7 human) for each bar. In all relevant panels, error bars indicate s.e.m., * indicates P<0.05 and ** P<0.01 as determined by Wilcoxon Rank test, and the scale bar indicates 10 μm.
Figure 2. Structure-activity relationship (SAR) analysis of “harmalogs” on beta cell proliferation
(a) Chemical structures of structural or functional harmine analogues (“harmalogs”). (b) and (c) Quantification of Ki67 labeling in rat (b) and human (c) beta cells in response to harmalogs. (d) An example of a Ki67+ “doublet” induced by INDY. The scale bar indicates 10 μm. (e−h) Dose-response curves for harmine and INDY in rat (e & g) and human (f & h) beta cell Ki67 labeling. In all relevant panels, error bars indicate s.e.m.; * indicates P<0.05 as determined by Wilcoxon Rank test. A minimum of 1000 beta cells was counted from four rat or four human donors for each graph. The scale bar indicates 10 μm.
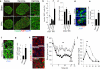
Figure 3. Calcineurin-NFAT-Dyrk1a signaling is implicated in harmine-Induced beta cell proliferation
(a) A model depicting the calcium (Ca++)−calmodulin (CAM)−calcineurin (CnA and CnB subunits)−NFAT−c-MYC pathway to beta cell proliferation. The sites of action of harmine, VIVIT and FK506 are shown in red. (b) Beta cell proliferation in dispersed human islets in response to harmine, in the presence and absence of VIVIT or FK506 (n = four human preparations). (c) Adenoviral Dyrk1a overexpression in human islets, visualized by immunoblot. Representative of experiments from three islet preparations. (d) Ad.DYRK1A overexpression and immunolabeling for DYRK1A in human beta cells. Representative of experiments from three human islet preparations. (e) Effects of Ad.DYRK1A overexpression on harmine- and INDY-induced human beta cell proliferation. A minimum of 1000 beta cells was counted from four donors for each bar. (f) Effects of Ad.sh_DYRK1A_ transduction in human islets. Representative of four human islet preparations. “Ad.Scr” indicates an adenovirus expressing scrambled shRNA. (g) An example of Ki67 immunolabeling in human beta cells transduced with Ad.sh_DYRK1A_. (h) Quantification of Ki67 in human beta cells transduced with Ad.sh_DYRK1A_. A minimum of 1000 beta cells was counted from five donors for each bar. (i) Effects of harmine (10 μM) or INDY (15 μM) treatment on NFAT4 translocation to the nucleus of human beta cells. Examples are shown in red arrows. Translocation of other NFATs is shown in Supplementary Figure 7. In all relevant panels, the scale bar indicates 10 μm, error bars indicate s.e.m., and * indicates P<0.05 as determined by Wilcoxon Rank test.
Figure 4. Effects of harmine in three in vivo models of beta cell replication and regeneration
(a−c) The PPX model: (a) Examples of Ki67 immunolabeling of beta cells in mice receiving a 60% partial pancreatectomy (PPX) or a sham operation, and treated either with vehicle or harmine for seven days. (b) Quantification of Ki67 labeling of beta cells in the four treatment groups, with four mice in each group. * indicates P<0.05 as determined by Wilcoxon Rank test. (c) Beta cell mass in the four groups following two weeks of harmine or vehicle, with four mice in each group. * indicates P<0.05 as determined by Wilcoxon Rank test. (d−h) The euglycemic model: (d) examples of BrdU labeling in human beta cells following seven days of harmine or vehicle (saline) treatment of NOD-SCID mice transplanted with human islets (_n_=three different donors) into the renal capsule. (e) Quantification of BrdU incorporation into transplanted human beta cells in the NOD-SCID mice in (d). * indicates P<0.05 as determined by Student's unpaired t-test. (f) An example of Ki67 immunolabeling of human beta cells in the same experiment as in d. (g) Quantification of Ki67 immunolabeling in human beta cells in d. (_n_=4). * indicates P<0.05 as determined by Student's unpaired t-test. (h) An example of TUNEL labeling of the three human islet grafts in d; DNAse treatment is a positive control; _n_=4. (i) The diabetic model: streptozotocin diabetic NOD-SCID mice transplanted with human islets, treated for 14 days with saline (grey lines, _n_=3) or harmine (black lines, _n_=4). Grafts were harvested at day 21 by unilateral nephrectomy (UNX). ** indicates P<0.01 for area under the curve as determined using unpaired Student's two-tailed _t_-text. (j) Intrapertioneal glucose tolerance test on Day 20 in the same mice as in i. ** as in i. In all relevant panels, bars show mean ± s.e.m. The scale bars indicate 50 μm.
Similar articles
- Novel factors modulating human β-cell proliferation.
Shirakawa J, Kulkarni RN. Shirakawa J, et al. Diabetes Obes Metab. 2016 Sep;18 Suppl 1(Suppl 1):71-7. doi: 10.1111/dom.12731. Diabetes Obes Metab. 2016. PMID: 27615134 Free PMC article. Review. - Development of Kinase-Selective, Harmine-Based DYRK1A Inhibitors that Induce Pancreatic Human β-Cell Proliferation.
Kumar K, Wang P, Sanchez R, Swartz EA, Stewart AF, DeVita RJ. Kumar K, et al. J Med Chem. 2018 Sep 13;61(17):7687-7699. doi: 10.1021/acs.jmedchem.8b00658. Epub 2018 Aug 21. J Med Chem. 2018. PMID: 30059217 Free PMC article. - Identification of harmine and β-carboline analogs from a high-throughput screen of an approved drug collection; profiling as differential inhibitors of DYRK1A and monoamine oxidase A and for in vitro and in vivo anti-cancer studies.
Tarpley M, Oladapo HO, Strepay D, Caligan TB, Chdid L, Shehata H, Roques JR, Thomas R, Laudeman CP, Onyenwoke RU, Darr DB, Williams KP. Tarpley M, et al. Eur J Pharm Sci. 2021 Jul 1;162:105821. doi: 10.1016/j.ejps.2021.105821. Epub 2021 Mar 27. Eur J Pharm Sci. 2021. PMID: 33781856 Free PMC article. - Combined Inhibition of DYRK1A, SMAD, and Trithorax Pathways Synergizes to Induce Robust Replication in Adult Human Beta Cells.
Wang P, Karakose E, Liu H, Swartz E, Ackeifi C, Zlatanic V, Wilson J, González BJ, Bender A, Takane KK, Ye L, Harb G, Pagliuca F, Homann D, Egli D, Argmann C, Scott DK, Garcia-Ocaña A, Stewart AF. Wang P, et al. Cell Metab. 2019 Mar 5;29(3):638-652.e5. doi: 10.1016/j.cmet.2018.12.005. Epub 2018 Dec 20. Cell Metab. 2019. PMID: 30581122 Free PMC article. - DYRK1A Inhibitors as Potential Therapeutics for β-Cell Regeneration for Diabetes.
Kumar K, Suebsuwong C, Wang P, Garcia-Ocana A, Stewart AF, DeVita RJ. Kumar K, et al. J Med Chem. 2021 Mar 25;64(6):2901-2922. doi: 10.1021/acs.jmedchem.0c02050. Epub 2021 Mar 8. J Med Chem. 2021. PMID: 33682417 Review.
Cited by
- Aberrant methylation underlies insulin gene expression in human insulinoma.
Karakose E, Wang H, Inabnet W, Thakker RV, Libutti S, Fernandez-Ranvier G, Suh H, Stevenson M, Kinoshita Y, Donovan M, Antipin Y, Li Y, Liu X, Jin F, Wang P, Uzilov A, Argmann C, Schadt EE, Stewart AF, Scott DK, Lambertini L. Karakose E, et al. Nat Commun. 2020 Oct 15;11(1):5210. doi: 10.1038/s41467-020-18839-1. Nat Commun. 2020. PMID: 33060578 Free PMC article. - Novel Approaches to Restore Pancreatic Beta-Cell Mass and Function.
Welters A, Lammert E. Welters A, et al. Handb Exp Pharmacol. 2022;274:439-465. doi: 10.1007/164_2021_474. Handb Exp Pharmacol. 2022. PMID: 34114119 Review. - A chemical with proven clinical safety rescues Down-syndrome-related phenotypes in through DYRK1A inhibition.
Kim H, Lee KS, Kim AK, Choi M, Choi K, Kang M, Chi SW, Lee MS, Lee JS, Lee SY, Song WJ, Yu K, Cho S. Kim H, et al. Dis Model Mech. 2016 Aug 1;9(8):839-48. doi: 10.1242/dmm.025668. Epub 2016 Jul 7. Dis Model Mech. 2016. PMID: 27483355 Free PMC article. - Novel factors modulating human β-cell proliferation.
Shirakawa J, Kulkarni RN. Shirakawa J, et al. Diabetes Obes Metab. 2016 Sep;18 Suppl 1(Suppl 1):71-7. doi: 10.1111/dom.12731. Diabetes Obes Metab. 2016. PMID: 27615134 Free PMC article. Review. - Metabolic control of adaptive β-cell proliferation by the protein deacetylase SIRT2.
Wortham M, Ramms B, Zeng C, Benthuysen JR, Sai S, Pollow DP, Liu F, Schlichting M, Harrington AR, Liu B, Prakash TP, Pirie EC, Zhu H, Baghdasarian S, Auwerx J, Shirihai OS, Sander M. Wortham M, et al. bioRxiv [Preprint]. 2024 Feb 28:2024.02.24.581864. doi: 10.1101/2024.02.24.581864. bioRxiv. 2024. PMID: 38464227 Free PMC article. Preprint.
References
- Kassem SA, Ariel I, Thornton PS, Scheimberg I, Glaser B. Beta cell proliferation and apoptosis in the developing normal human pancreas and in hyperinsulinism of infancy. Diabetes. 2000;49:1325–1333. - PubMed
- Kohler CU, Olewinski M, Tannapfel A, Schmidt WE, Fritsch H, Meier JJ. Cell cycle control of beta cell replication in the prenatal and postnatal human pancreas. Am J Physiol Endocrinol Metab. 2011;300:E221–230. - PubMed
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta cell deficit and increased beta cell apoptosis in humans with diabetes. Diabetes. 2003;52:102–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R-01 DK55023/DK/NIDDK NIH HHS/United States
- R-01 DK077096/DK/NIDDK NIH HHS/United States
- R-01 DK065149/DK/NIDDK NIH HHS/United States
- R01 DK067351/DK/NIDDK NIH HHS/United States
- R01 DK055023/DK/NIDDK NIH HHS/United States
- U01 DK089538/DK/NIDDK NIH HHS/United States
- R01 DK065149/DK/NIDDK NIH HHS/United States
- R01 DK105015/DK/NIDDK NIH HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- P60 DK020541/DK/NIDDK NIH HHS/United States
- R01 DK077096/DK/NIDDK NIH HHS/United States
- U-01 DK089538/DK/NIDDK NIH HHS/United States
- R-01 DK067351/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous