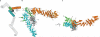Atomic structure of the Y complex of the nuclear pore - PubMed (original) (raw)
. 2015 May;22(5):425-431.
doi: 10.1038/nsmb.2998. Epub 2015 Mar 30.
Affiliations
- PMID: 25822992
- PMCID: PMC4424061
- DOI: 10.1038/nsmb.2998
Atomic structure of the Y complex of the nuclear pore
Kotaro Kelley et al. Nat Struct Mol Biol. 2015 May.
Abstract
The nuclear pore complex (NPC) is the principal gateway for transport into and out of the nucleus. Selectivity is achieved through the hydrogel-like core of the NPC. The structural integrity of the NPC depends on ~15 architectural proteins, which are organized in distinct subcomplexes to form the >40-MDa ring-like structure. Here we present the 4.1-Å crystal structure of a heterotetrameric core element ('hub') of the Y complex, the essential NPC building block, from Myceliophthora thermophila. Using the hub structure together with known Y-complex fragments, we built the entire ~0.5-MDa Y complex. Our data reveal that the conserved core of the Y complex has six rather than seven members. Evolutionarily distant Y-complex assemblies share a conserved core that is very similar in shape and dimension, thus suggesting that there are closely related architectural codes for constructing the NPC in all eukaryotes.
Figures
Figure 1. Structure of the Myceliophthora thermophila Y-complex hub at 4.1 Å resolution
(a) Schematic of the Y-complex. Regions included in the crystallization construct are colored, other Y-complex regions in grey. Elements of ACE1 fold proteins are indicated: T – tail and C – crown flank the central trunk element. (b) The hub structure is colored as follows: Nup85 (orange), Nup120 (green), Nup145C (cyan), and the Sec13 β-propeller (grey). N and C termini are indicated for the helical proteins. Helices are numbered according to previously solved S. cerevisiae fragments,,. Numbers that include a letter modifier indicate helical elements not present in S. cerevisiae. (c) Top-down view of the hub. The N terminus of Nup145C is not indicated because it is obscured by the 85° rotation.
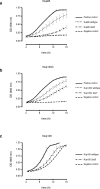
Figure 2. Fitness analysis of hub interactions
(a) Growth curves of _NUP85_Δ strains carrying NUP85:URA3 and either empty pRS315 (negative control), Nup85 wildtype, or Nup85 Δα30 grown in the presence of 5-FOA. The positive control is the _NUP85_Δ strain carrying NUP85/URA3 and empty pRS315 grown in the absence of 5-FOA. (b) Growth curves of _NUP145_Δ strains carrying NUP145/URA3 and either empty pRS315 (negative control), Nup145C wildtype, or Nup145C Δα27 grown in the presence of 5-FOA. The positive control is the _NUP145_Δ strain carrying NUP145/URA3 and empty pRS315 grown in the absence of 5-FOA. (c) The growth curves of _NUP120_Δ strains carrying YClac33 empty vector (negative control), Nup120 wildtype, or Nup120 Δα30 grown in YPD. Four technical replicates (_n_=4 OD measurements) for each of three biological replicates (_n_=3), from separate colonies, were performed at 30 °C for all experiments. All error bars are standard deviation of the mean (s.e.m.).
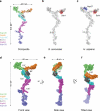
Figure 3. Composite high-resolution structure of the Y-complex
(a) Composite, hexameric Y-complex core constructed from the hub structure (Fig. 1) combined with previously published X-ray crystal structure fragments. Within Nup84, 4 helices were modeled computationally. (b) Composite S. cerevisiae Y-complex based on A), with S. cerevisiae sequences threaded onto existing homologous structures. Compared to the universally conserved hexameric Y-complex core shown in A), Seh1 (red) is an additional component found in many organisms, including yeast. (c) Composite H. sapiens Y-complex with H. sapiens sequences threaded onto existing homologous structures. Nup37 (blue) is another Y-complex component only found in a subset of eukaryotes, including humans. (d) Space filling surface view of the composite, hexameric Y-complex viewed from the front. (e) Side view. (f) Tilted view.
Figure 4. Comparison of the X-ray based, composite Y-complex structure with published 3-D EM reconstruction structures
(a,b) Electron density envelope around the composite H. sapiens Y-complex calculated for 35 Å resolution from front (a) or top (b) view. (c,d) 3-D EM reconstruction of the H. sapiens Y-complex with an overlay of the composite model, fitted computationally from front (c) or top (d) view. (e,f)Electron density envelope around the composite S. cerevisiae Y-complex calculated at 33 Å resolution from front (e) or top (f) view. (g,h) 3-D EM reconstruction of the S. cerevisiae Y-complex with an overlay of the composite model, fitted computationally from front (g) or top (h) view.
Figure 5. Flexibility of the Y-complex
Experimentally observed hinge regions of the Y-complex are denoted by dashed lines. (a) ref. . (b–c) ref. . (d–e), refs. -,,. (f) ref. .
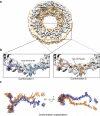
Figure 6. Fitting of the composite H. sapiens Y-complex into the cryo-ET map of the entire NPC
(a) Consensus map calculated from the cryo-ET map of the human NPC (EMD code: 2444). The cytoplasmic ring density is highlighted in grey for clarity. (b) The top scoring fit of the composite H. sapiens Y-complex (conformation 1) and the second top scoring fit (conformation 2) are depicted. (c) A superposition of the two fits from (b) shows that they are related by a ~20° rotation about the Y-complex hub, and substantial bending of the long stack.
Similar articles
- Utilizing the Dyn2 dimerization-zipper as a tool to probe NPC structure and function.
Flemming D, Stelter P, Hurt E. Flemming D, et al. Methods Cell Biol. 2014;122:99-115. doi: 10.1016/B978-0-12-417160-2.00005-9. Methods Cell Biol. 2014. PMID: 24857727 - A nanobody suite for yeast scaffold nucleoporins provides details of the nuclear pore complex structure.
Nordeen SA, Andersen KR, Knockenhauer KE, Ingram JR, Ploegh HL, Schwartz TU. Nordeen SA, et al. Nat Commun. 2020 Dec 2;11(1):6179. doi: 10.1038/s41467-020-19884-6. Nat Commun. 2020. PMID: 33268786 Free PMC article. - Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
Whittle JRR, Schwartz TU. Whittle JRR, et al. J Biol Chem. 2009 Oct 9;284(41):28442-28452. doi: 10.1074/jbc.M109.023580. Epub 2009 Aug 11. J Biol Chem. 2009. PMID: 19674973 Free PMC article. - The diverse roles of the Nup93/Nic96 complex proteins - structural scaffolds of the nuclear pore complex with additional cellular functions.
Vollmer B, Antonin W. Vollmer B, et al. Biol Chem. 2014 May;395(5):515-28. doi: 10.1515/hsz-2013-0285. Biol Chem. 2014. PMID: 24572986 Review. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
Cited by
- Molecular determinants of large cargo transport into the nucleus.
Paci G, Zheng T, Caria J, Zilman A, Lemke EA. Paci G, et al. Elife. 2020 Jul 21;9:e55963. doi: 10.7554/eLife.55963. Elife. 2020. PMID: 32692309 Free PMC article. - Identification and localization of Nup170 in the microsporidian Nosema bombycis.
Shang R, Zhu F, Li Y, He P, Qi J, Chen Y, Sun F, Zhang Y, Wang Q, Shen Z. Shang R, et al. Parasitol Res. 2021 Jun;120(6):2125-2134. doi: 10.1007/s00436-021-07129-4. Epub 2021 Mar 26. Parasitol Res. 2021. PMID: 33768334 - Assembly principle of a membrane-anchored nuclear pore basket scaffold.
Cibulka J, Bisaccia F, Radisavljević K, Gudino Carrillo RM, Köhler A. Cibulka J, et al. Sci Adv. 2022 Feb 11;8(6):eabl6863. doi: 10.1126/sciadv.abl6863. Epub 2022 Feb 11. Sci Adv. 2022. PMID: 35148185 Free PMC article. - Breaking the Y.
Holzer G, Antonin W. Holzer G, et al. PLoS Genet. 2019 May 23;15(5):e1008109. doi: 10.1371/journal.pgen.1008109. eCollection 2019 May. PLoS Genet. 2019. PMID: 31120884 Free PMC article. No abstract available. - Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article.
References
- Strambio-de-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:490–501. - PubMed
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell. 1998;1:223–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM077537/GM/NIGMS NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
- R01GM77537/GM/NIGMS NIH HHS/United States
- T32GM007287/GM/NIGMS NIH HHS/United States
- F31 GM115068/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases