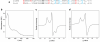Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink - PubMed (original) (raw)
Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink
Kelsey R Schramma et al. Nat Chem. 2015 May.
Abstract
Streptococcal bacteria use peptide signals as a means of intraspecies communication. These peptides can contain unusual post-translational modifications, providing opportunities for expanding our understanding of nature's chemical and biosynthetic repertoires. Here, we have combined tools from natural products discovery and mechanistic enzymology to elucidate the structure and biosynthesis of streptide, a streptococcal macrocyclic peptide. We show that streptide bears an unprecedented post-translational modification involving a covalent linkage between two unactivated carbons within the side chains of lysine and tryptophan. The biosynthesis of streptide was addressed by genetic and biochemical studies. The former implicated a new SPASM-domain-containing radical SAM enzyme StrB, while the latter revealed that StrB contains two [4Fe-4S] clusters and installs the unusual lysine-to-tryptophan crosslink in a single step. By intramolecularly stitching together the side chains of lysine and tryptophan, StrB provides a new route for biosynthesizing macrocyclic peptides.
Figures
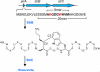
Figure 1. Biosynthetic gene cluster and structure of streptide
a, The str cluster consists of a precursor peptide (strA), a radical SAM gene (strB), and a putative transporter (strC). It is activated by the shp/rgg quorum sensing system, which encodes a peptide hormone (shp) and a transcriptional regulator (rgg)14. b, Structural elucidation of streptide by NMR. Key correlations used to solve the structure are indicated. c, Structural and stereochemical assignment of streptide. d, Computational model of the three-dimensional structure of streptide calculated using CYANA and NMR NOESY constraints. e, HR-HPLC-MS analysis of the extracts of wt S. thermophilus (red trace) and a strB::ery mutant (black trace). Shown is the extracted ion intensity for streptide (m/z 989.4879). The starred peak corresponds to streptide.
Figure 2. Proposed biosynthesis for streptide
StrA encodes a 30mer substrate. In our model, StrB installs the Lys-to-Trp crosslink, while StrC, possibly in concert with other protease(s), secretes the mature 9mer streptide. A 9mer, 20mer, and the full-length 30mer StrA, synthesized in this study, are shown. The 9mer sequence of streptide is shown in bold; the K and W residues involved in the cyclization are shown in red.
Figure 3. StrB contains two [4Fe-4S] clusters
a, Alignment of the SPASM motif of StrB with that of anSME. Cys residues that bind the AuxI and AuxII clusters (in anSME) are shown in red and blue, respectively. StrB lacks a key Cys ligand to the AuxI cluster. b, UV-vis spectrum of reconstituted StrB showing a 320 nm shoulder and a 395 nm feature, characteristic of [4Fe-4S] clusters. c, X-band CW EPR spectrum of reconstituted StrB yielding gx, gy, and gz of 1.86, 1.94, and 2.05, respectively. d, EPR spectrum of C409A/C415A-StrB with gII and g of 2.06 and 1.93 typical for the SAM-cleaving [4Fe-4S]+ cluster.
Figure 4. StrB catalyzes Lys-to-Trp crosslink formation in StrA
a, Qtof-HPLC-MS assay for 30mer product formation. Activity assays were carried out in the absence of 30mer substrate (gray trace), reductant (black trace), SAM (blue trace), StrB (green trace), or containing all components (red trace). The 30mer substrate (S) and product (P) peaks are marked. The traces have been offset in both axes for clarity. Inset, HR-ESI-MS spectrum of product 30mer ([M+2H]2+ calc 1656.29144). b, Overlaid TOCSY NMR spectra of StrA (blue) and the product 30mer (red). The substrate contains two unmodified Trp residues (WC and WN), while the product spectrum reveals a C-terminal Trp (WC) and the modified Trp lacking a 1H at the indole-C7 (Wm). The peaks have been assigned in the overlaid 1H traces. See Supplementary Fig. 15 for the upstream region. c, HR-MS/MS spectrum of product 30mer. The observed b and y ions are shown in the inset. Fragmentation at all peptide bonds is observed except for those in the KGDGW sequence, consistent with macrocyclization within this motif (see Supplementary Table 5).
Figure 5. Mechanistic model for Lys-to-Trp crosslink formation catalysed by StrB
The FeS clusters in red and blue correspond to the SAM-cleaving active site cluster and the auxiliary cluster, respectively. Reductive activation of SAM leads to formation of 5′-dA•, which abstracts a Lys β-hydrogen. The radical thus formed reacts with the indole side chain to create the Lys-to-Trp crosslink and an indolyl radical. Deprotonation and rearomatization with concomitant reduction of the auxiliary Fe-S cluster completes the synthesis of crosslinked StrA.
Similar articles
- Total Synthesis and Stereochemical Assignment of Streptide.
Isley NA, Endo Y, Wu ZC, Covington BC, Bushin LB, Seyedsayamdost MR, Boger DL. Isley NA, et al. J Am Chem Soc. 2019 Oct 30;141(43):17361-17369. doi: 10.1021/jacs.9b09067. Epub 2019 Oct 17. J Am Chem Soc. 2019. PMID: 31577142 Free PMC article. - Lysine-Tryptophan-Crosslinked Peptides Produced by Radical SAM Enzymes in Pathogenic Streptococci.
Schramma KR, Seyedsayamdost MR. Schramma KR, et al. ACS Chem Biol. 2017 Apr 21;12(4):922-927. doi: 10.1021/acschembio.6b01069. Epub 2017 Mar 3. ACS Chem Biol. 2017. PMID: 28191919 - Insights into the catalysis of a lysine-tryptophan bond in bacterial peptides by a SPASM domain radical _S_-adenosylmethionine (SAM) peptide cyclase.
Benjdia A, Decamps L, Guillot A, Kubiak X, Ruffié P, Sandström C, Berteau O. Benjdia A, et al. J Biol Chem. 2017 Jun 30;292(26):10835-10844. doi: 10.1074/jbc.M117.783464. Epub 2017 May 5. J Biol Chem. 2017. PMID: 28476884 Free PMC article. - Tryptophan tryptophylquinone biosynthesis: a radical approach to posttranslational modification.
Davidson VL, Liu A. Davidson VL, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1299-305. doi: 10.1016/j.bbapap.2012.01.008. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22314272 Free PMC article. Review. - Construction and screening of vast libraries of natural product-like macrocyclic peptides using in vitro display technologies.
Bashiruddin NK, Suga H. Bashiruddin NK, et al. Curr Opin Chem Biol. 2015 Feb;24:131-8. doi: 10.1016/j.cbpa.2014.11.011. Epub 2014 Dec 5. Curr Opin Chem Biol. 2015. PMID: 25483262 Review.
Cited by
- Sequence-function space of radical SAM cyclophane synthases reveal conserved active site residues that influence substrate specificity.
Phan CS, Morinaka BI. Phan CS, et al. RSC Chem Biol. 2024 Oct 23;5(12):1195-200. doi: 10.1039/d4cb00227j. Online ahead of print. RSC Chem Biol. 2024. PMID: 39464308 Free PMC article. - Use of Rgg quorum-sensing machinery to create an innovative recombinant protein expression system in Streptococcus thermophilus.
Gardan R, Honvo-Houeto E, Mézange C, Maillot NJ, Balvay A, Rabot S, Bermúdez-Humarán LG, Langella P, Monnet V, Juillard V. Gardan R, et al. Microbiology (Reading). 2024 Sep;170(9):001487. doi: 10.1099/mic.0.001487. Microbiology (Reading). 2024. PMID: 39302176 Free PMC article. - The Metabolic Potential of the Human Lung Microbiome.
Semmler F, Regis Belisário-Ferrari M, Kulosa M, Kaysser L. Semmler F, et al. Microorganisms. 2024 Jul 17;12(7):1448. doi: 10.3390/microorganisms12071448. Microorganisms. 2024. PMID: 39065215 Free PMC article. - Genome Mining for New Enzyme Chemistry.
Nguyen DT, Mitchell DA, van der Donk WA. Nguyen DT, et al. ACS Catal. 2024 Mar 12;14(7):4536-4553. doi: 10.1021/acscatal.3c06322. eCollection 2024 Apr 5. ACS Catal. 2024. PMID: 38601780 Free PMC article. Review. - A multicomponent reaction for modular assembly of indole-fused heterocycles.
Li J, Ni H, Zhang W, Lai Z, Jin H, Zeng L, Cui S. Li J, et al. Chem Sci. 2024 Mar 4;15(14):5211-5217. doi: 10.1039/d4sc00522h. eCollection 2024 Apr 3. Chem Sci. 2024. PMID: 38577354 Free PMC article.
References
- Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature. 2000;407:215–218. - PubMed
- Duquesne S, et al. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem. Biol. 2007;14:793–803. - PubMed
- Dorenbos R, et al. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 2002;277:16682–16688. - PubMed
- Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew. Chem. Int. Ed. 2008;47:7756–7759. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources