Chromatin plasticity in response to DNA damage: The shape of things to come - PubMed (original) (raw)
Review
Chromatin plasticity in response to DNA damage: The shape of things to come
Salomé Adam et al. DNA Repair (Amst). 2015 Aug.
Abstract
DNA damage poses a major threat to cell function and viability by compromising both genome and epigenome integrity. The DNA damage response indeed operates in the context of chromatin and relies on dynamic changes in chromatin organization. Here, we review the molecular bases of chromatin alterations in response to DNA damage, focusing on core histone mobilization in mammalian cells. Building on our current view of nucleosome dynamics in response to DNA damage, we highlight open challenges and avenues for future development. In particular, we discuss the different levels of regulation of chromatin plasticity during the DNA damage response and their potential impact on cell function and epigenome maintenance.
Keywords: Chromatin dynamics; Chromatin remodelers; DNA damage repair; Epigenome maintenance; Histone chaperones; Histone variants.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures
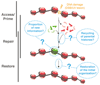
Fig.1. Histone dynamics in response to DNA damage: the issue of epigenome maintenance.
DNA damage (yellow star) elicits important chromatin rearrangements, including a loss of parental information (red) at the damage site due to the mobilization of pre-existing histones, and the incorporation of new information (green) with DNA damage-responsive PTMs, histone variant exchange and deposition of newly synthesized histones. The resulting pattern of histone variants and associated PTMs is likely to differ substantially from the original one. Future challenges in the field (open issues in blue) will be to determine to which extent the original information is diluted and whether or not the pre-existing chromatin landscape is ultimately faithfully restored after genotoxic stress, by parental histone recycling, histone variant exchange, active erasure of DNA damage-associated PTMs, and/or transmission of parental marks to newly deposited histones.
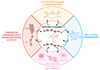
Fig.2. Cross-talks between histone dynamics in damaged chromatin and cellular functions.
The exchange of parental histones (red) with histones carrying new information (green) in response to DNA damage will impose - at least transient - changes in chromatin structure and function. Future work should closely examine to which extent these dynamics impact DNA metabolic activities (such as DNA repair and transcription) and cellular functions (proliferation and differentiation), and assess their consequences on cell identity. Conversely, it needs to be further explored how the initial chromatin organization, DNA damage type, cell cycle stage and cell state may regulate DNA damage-induced histone dynamics.
Similar articles
- Chromatin assembly: a basic recipe with various flavours.
Polo SE, Almouzni G. Polo SE, et al. Curr Opin Genet Dev. 2006 Apr;16(2):104-11. doi: 10.1016/j.gde.2006.02.011. Epub 2006 Feb 28. Curr Opin Genet Dev. 2006. PMID: 16504499 Review. - Histone core modifications regulating nucleosome structure and dynamics.
Tessarz P, Kouzarides T. Tessarz P, et al. Nat Rev Mol Cell Biol. 2014 Nov;15(11):703-8. doi: 10.1038/nrm3890. Epub 2014 Oct 15. Nat Rev Mol Cell Biol. 2014. PMID: 25315270 Review. - Histone chaperones: assisting histone traffic and nucleosome dynamics.
Gurard-Levin ZA, Quivy JP, Almouzni G. Gurard-Levin ZA, et al. Annu Rev Biochem. 2014;83:487-517. doi: 10.1146/annurev-biochem-060713-035536. Annu Rev Biochem. 2014. PMID: 24905786 Review. - Chromatin response to DNA double-strand break damage.
Bao Y. Bao Y. Epigenomics. 2011 Jun;3(3):307-21. doi: 10.2217/epi.11.14. Epigenomics. 2011. PMID: 22122340 Review.
Cited by
- Regulation of Nuclear Mechanics and the Impact on DNA Damage.
Dos Santos Á, Toseland CP. Dos Santos Á, et al. Int J Mol Sci. 2021 Mar 20;22(6):3178. doi: 10.3390/ijms22063178. Int J Mol Sci. 2021. PMID: 33804722 Free PMC article. Review. - Chromatin Dynamics in Genome Stability: Roles in Suppressing Endogenous DNA Damage and Facilitating DNA Repair.
Nair N, Shoaib M, Sørensen CS. Nair N, et al. Int J Mol Sci. 2017 Jul 10;18(7):1486. doi: 10.3390/ijms18071486. Int J Mol Sci. 2017. PMID: 28698521 Free PMC article. Review. - Design, synthesis, and characterization of nucleosomes containing site-specific DNA damage.
Taylor JS. Taylor JS. DNA Repair (Amst). 2015 Dec;36:59-67. doi: 10.1016/j.dnarep.2015.09.025. Epub 2015 Oct 19. DNA Repair (Amst). 2015. PMID: 26493358 Free PMC article. Review. - Super-Resolution Localization Microscopy of γ-H2AX and Heterochromatin after Folate Deficiency.
Bach M, Savini C, Krufczik M, Cremer C, Rösl F, Hausmann M. Bach M, et al. Int J Mol Sci. 2017 Aug 8;18(8):1726. doi: 10.3390/ijms18081726. Int J Mol Sci. 2017. PMID: 28786938 Free PMC article. - Chromosome territory relocation paradigm during DNA damage response: Some insights from molecular biology to physics.
Fatakia SN, Kulashreshtha M, Mehta IS, Rao BJ. Fatakia SN, et al. Nucleus. 2017 Sep 3;8(5):449-460. doi: 10.1080/19491034.2017.1313938. Epub 2017 Jun 22. Nucleus. 2017. PMID: 28640660 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources