Global, in situ, site-specific analysis of protein S-sulfenylation - PubMed (original) (raw)
Global, in situ, site-specific analysis of protein S-sulfenylation
Jing Yang et al. Nat Protoc. 2015 Jul.
Abstract
Protein S-sulfenylation is the reversible oxidative modification of cysteine thiol groups to form cysteine S-sulfenic acids. Mapping the specific sites of protein S-sulfenylation onto complex proteomes is crucial to understanding the molecular mechanisms controlling redox signaling and regulation. This protocol describes global, in situ, site-specific analysis of protein S-sulfenylation using sulfenic acid-specific chemical probes and mass spectrometry (MS)-based proteomics. The major steps in this protocol are as follows: (i) optimization of conditions for selective labeling of cysteine S-sulfenic acids in intact cells with the commercially available dimedone-based probe, DYn-2; (ii) tagging the modified cysteines with a functionalized biotin reagent containing a cleavable linker via Cu(I)-catalyzed azide-alkyne cycloaddition reaction; (iii) enrichment of the biotin-tagged tryptic peptides with streptavidin; (iv) liquid chromatography-tandem MS (LC-MS/MS)-based shotgun proteomics; and (v) computational data analysis. We also outline strategies for quantitative analysis of this modification in cells responding to redox perturbations and discuss special issues pertaining to experimental design of thiol redox studies. Our chemoproteomic platform should be broadly applicable to the investigation of other bio-orthogonal chemically engineered post-translational modifications. The entire analysis protocol takes ∼1 week to complete.
Conflict of interest statement
COMPETING FINANCIAL INTERSESTS
The authors declare no competing financial interests.
Figures
Figure 1
Selective labeling and detection of protein S-sulphenic acid in cells. (a) Cyclohexanedione group of DYn-2 probe selectively reacts with protein sulphenic acid modifications in cells. To the best of our knowledge, this probe has no reactivity toward other functional groups in cells. Moreover, this probe does not change cell viability and redox balance. (b) Alkyne group of DYn-2-labeled proteins can be conjugated to azide-bearing tags (e.g. fluorescent or biotin tag) via click chemistry and detected by immunofluorescence, western blotting (WB), or proteomics.
Figure 2
Two workflows for proteome-wide identification of targeted sites of alkyne tagged probes. (a) Protein capture. Proteome labeled with an alkynyl probe is first conjugated with an azido biotin with cleavable linker via CuAAC chemistry. The biotinylated proteins are then enriched with streptavidin beads and subjected to sequential on-beads tryptic digestion and released by an enzymatic or chemical cleavage mechanism. The resulting peptides are analyzed by mass spectrometry based proteomics for site-localization. (b) Peptide capture, as described in the Procedure. Proteome labeled with an alkynyl probe is first digested into tryptic peptides and then conjugated with an azido biotin with photocleavable linker via CuAAC chemistry. The biotinylated peptides are then enriched with streptavidin beads and released by an enzymatic or chemical cleavage mechanism. Sequential LC-MS analysis and informatics pipeline provide identification of labeled peptides, proteins and their sites of modification.
Figure 3
Identification of Cys238 of NAA15 as the target of S-sulphenylation in RKO cells. (a) Fully annotated HCD MS/MS spectrum with diagnostic fragment ions (zoom window) of a DYn-2-triazo-hexanoic-acid modified peptide from S-sulphenylated NAA15. The highlighted cysteine in the peptide sequence represents the S-sulphenylated site (Cys238) of NAA15. (b) CID MS/MS spectrum of the same peptide.
Figure 4
Identification of Cys152 (upper) and Cys156 (lower) of GAPDH as the targets of S-sulphenylation in RKO cells. Fully annotated HCD MS/MS spectra of the same peptide sequence with different sites of DYn-2-triazo-hexanoic-acid modification are shown. The cysteine in purple color in the peptide sequence represents the S-sulphenylated site.
Figure 5
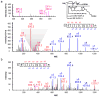
Quantitative S-sulphenylome analysis. (a) The proteome-wide and site-specific changes in S-sulfenylation in intact RKO cells in response to exogenous H2O2 (500 μM, 5 min), where nine S-sulfenylated cysteines on the FASN protein are shown in red, and all other cysteines by white symbols. The relative change in protein S-sulphenylation between two conditions is determined by the ratio of extracted ion current peak intensities for heavy and light isotope-labeled DYn-2-tagged peptides, using the MS1 filtering function of Skyline software. Ratios are displayed on a log2 scale on the y axis. (b) Extracted ion chromatograms (XIC) for nine S-sulfenylated peptides/sites from FASN are shown, with the profiles for light- and heavy- labeled peptides in red and blue, respectively. Light labels correspond to untreated cells; heavy labels correspond to H2O2-treated cells. The mean calculated experimental ratios from at least four replicates are displayed below the individual chromatogram, respectively. Figure reproduced from ref..
Similar articles
- Proteome-Wide Analysis of Cysteine S-Sulfenylation Using a Benzothiazine-Based Probe.
Fu L, Liu K, Ferreira RB, Carroll KS, Yang J. Fu L, et al. Curr Protoc Protein Sci. 2019 Feb;95(1):e76. doi: 10.1002/cpps.76. Epub 2018 Oct 12. Curr Protoc Protein Sci. 2019. PMID: 30312022 Free PMC article. Review. - Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation.
Shi Y, Carroll KS. Shi Y, et al. Acc Chem Res. 2020 Jan 21;53(1):20-31. doi: 10.1021/acs.accounts.9b00562. Epub 2019 Dec 23. Acc Chem Res. 2020. PMID: 31869209 Free PMC article. Review. - Cysteine-Mediated Redox Regulation of Cell Signaling in Chondrocytes Stimulated With Fibronectin Fragments.
Wood ST, Long DL, Reisz JA, Yammani RR, Burke EA, Klomsiri C, Poole LB, Furdui CM, Loeser RF. Wood ST, et al. Arthritis Rheumatol. 2016 Jan;68(1):117-26. doi: 10.1002/art.39326. Arthritis Rheumatol. 2016. PMID: 26314228 Free PMC article. - Isotope-coded chemical reporter and acid-cleavable affinity reagents for monitoring protein sulfenic acids.
Truong TH, Garcia FJ, Seo YH, Carroll KS. Truong TH, et al. Bioorg Med Chem Lett. 2011 Sep 1;21(17):5015-20. doi: 10.1016/j.bmcl.2011.04.115. Epub 2011 May 3. Bioorg Med Chem Lett. 2011. PMID: 21601453 - Identification of sulfenylation patterns in trophozoite stage Plasmodium falciparum using a non-dimedone based probe.
Schipper S, Wu H, Furdui CM, Poole LB, Delahunty CM, Park R, Yates JR 3rd, Becker K, Przyborski JM. Schipper S, et al. Mol Biochem Parasitol. 2021 Mar;242:111362. doi: 10.1016/j.molbiopara.2021.111362. Epub 2021 Jan 26. Mol Biochem Parasitol. 2021. PMID: 33513391 Free PMC article.
Cited by
- Dynamic redox balance directs the oocyte-to-embryo transition via developmentally controlled reactive cysteine changes.
Petrova B, Liu K, Tian C, Kitaoka M, Freinkman E, Yang J, Orr-Weaver TL. Petrova B, et al. Proc Natl Acad Sci U S A. 2018 Aug 21;115(34):E7978-E7986. doi: 10.1073/pnas.1807918115. Epub 2018 Aug 6. Proc Natl Acad Sci U S A. 2018. PMID: 30082411 Free PMC article. - Proteome-Wide Analysis of Cysteine S-Sulfenylation Using a Benzothiazine-Based Probe.
Fu L, Liu K, Ferreira RB, Carroll KS, Yang J. Fu L, et al. Curr Protoc Protein Sci. 2019 Feb;95(1):e76. doi: 10.1002/cpps.76. Epub 2018 Oct 12. Curr Protoc Protein Sci. 2019. PMID: 30312022 Free PMC article. Review. - Triphenylphosphonium-Derived Protein Sulfenic Acid Trapping Agents: Synthesis, Reactivity, and Effect on Mitochondrial Function.
Li Z, Forshaw TE, Holmila RJ, Vance SA, Wu H, Poole LB, Furdui CM, King SB. Li Z, et al. Chem Res Toxicol. 2019 Mar 18;32(3):526-534. doi: 10.1021/acs.chemrestox.8b00385. Epub 2019 Mar 4. Chem Res Toxicol. 2019. PMID: 30784263 Free PMC article. - Protein cysteine oxidation in redox signaling: Caveats on sulfenic acid detection and quantification.
Forman HJ, Davies MJ, Krämer AC, Miotto G, Zaccarin M, Zhang H, Ursini F. Forman HJ, et al. Arch Biochem Biophys. 2017 Mar 1;617:26-37. doi: 10.1016/j.abb.2016.09.013. Epub 2016 Sep 28. Arch Biochem Biophys. 2017. PMID: 27693037 Free PMC article. Review. - NOX1-dependent redox signaling potentiates colonic stem cell proliferation to adapt to the intestinal microbiota by linking EGFR and TLR activation.
van der Post S, Birchenough GMH, Held JM. van der Post S, et al. Cell Rep. 2021 Apr 6;35(1):108949. doi: 10.1016/j.celrep.2021.108949. Cell Rep. 2021. PMID: 33826887 Free PMC article.
References
- Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. - PubMed
- Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 CA159988/CA/NCI NIH HHS/United States
- U24CA159988/CA/NCI NIH HHS/United States
- R01GM102187/GM/NIGMS NIH HHS/United States
- R01 HD064727/HD/NICHD NIH HHS/United States
- R01 CA174864/CA/NCI NIH HHS/United States
- R01 GM102187/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources