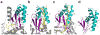A prevalent peptide-binding domain guides ribosomal natural product biosynthesis - PubMed (original) (raw)
A prevalent peptide-binding domain guides ribosomal natural product biosynthesis
Brandon J Burkhart et al. Nat Chem Biol. 2015 Aug.
Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a rapidly growing class of natural products. RiPP precursor peptides can undergo extensive enzymatic tailoring to yield structurally and functionally diverse products, and their biosynthetic logic makes them attractive bioengineering targets. Recent work suggests that unrelated RiPP-modifying enzymes contain structurally similar precursor peptide-binding domains. Using profile hidden Markov model comparisons, we discovered related and previously unrecognized peptide-binding domains in proteins spanning the majority of known prokaryotic RiPP classes, and we named this conserved domain the RiPP precursor peptide recognition element (RRE). Through binding studies we verified RRE's roles for three distinct RiPP classes: linear azole-containing peptides, thiopeptides and lasso peptides. Because numerous RiPP biosynthetic enzymes act on peptide substrates, our findings have powerful predictive value as to which protein(s) drive substrate binding, thereby laying a foundation for further characterization of RiPP biosynthetic pathways and the rational engineering of new peptide-binding activities.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
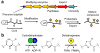
Figure 1. Overview of RiPP biosynthesis and the TOMM subclass
(a) A generic RiPP biosynthetic gene cluster is displayed. The precursor peptide is composed of a leader peptide (LP) and core. While the LP contains binding motifs for the modifying enzymes, the core contains residues that undergo enzymatic processing to diverse functional groups. After removal of the LP and any additional tailoring processes, the mature RiPP is exported. (b) One RiPP biosynthetic class, the thiazole/oxazole-modified microcins (TOMMs), installs azoline/azole heterocycles. TOMMs arise from the action of an ATP-dependent cyclodehydratase (C and D proteins) and a flavin mononucleotide (FMN)-dependent dehydrogenase (B protein). X = S or O; R = H or CH3.
Figure 2. Structural comparison of four RiPP modifying enzymes
Shown are structurally equivalent sections of RiPP biosynthetic proteins involved in the biosynthesis of (a) the Trojan horse antibiotic microcin C7 (MccB, an adenylating enzyme [PDB entry 3H9J]), (b) antitumor cyanobactins (LynD, a cyclodehydratase [4V1T]), (c) the lantibiotic nisin (NisB, dehydratase [4WD9]), and (d) the bacterial dehydrogenase cofactor PQQ (PqqD, rSAM-associated [3G2B]). The purple β-sheets and cyan α-helices constitute a conserved
R
iPP precursor peptide
r
ecognition
e
lement (RRE). In (a–c), the precursor peptide is shown in yellow stick format. Dashed lines indicate missing electron density.
Figure 3. RREs are present in diverse RiPP biosynthetic proteins
RREs were found in a myriad of RiPP biosynthetic proteins using HHpred to search for PqqD homology (thick black bars). Solid lines represent protein sequences with N/_C_-termini labeled as “N” and “C.” Colored sections indicate the annotations for the conserved domains identified by the Conserved Domain Database. Asterisks denote RRE assignments based solely on HHpred findings. Abbreviations: LAP, linear azole-containing peptide; PQQ, pyrroloquinoline quinone; rSAM, radical SAM. More details can be found in Supplementary Table 3.
Similar articles
- New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products.
Ortega MA, van der Donk WA. Ortega MA, et al. Cell Chem Biol. 2016 Jan 21;23(1):31-44. doi: 10.1016/j.chembiol.2015.11.012. Cell Chem Biol. 2016. PMID: 26933734 Free PMC article. Review. - The B1 Protein Guides the Biosynthesis of a Lasso Peptide.
Zhu S, Fage CD, Hegemann JD, Mielcarek A, Yan D, Linne U, Marahiel MA. Zhu S, et al. Sci Rep. 2016 Oct 18;6:35604. doi: 10.1038/srep35604. Sci Rep. 2016. PMID: 27752134 Free PMC article. - Ribosomal Natural Products, Tailored To Fit.
Funk MA, van der Donk WA. Funk MA, et al. Acc Chem Res. 2017 Jul 18;50(7):1577-1586. doi: 10.1021/acs.accounts.7b00175. Epub 2017 Jul 6. Acc Chem Res. 2017. PMID: 28682627 Free PMC article. - Expansion of RiPP biosynthetic space through integration of pan-genomics and machine learning uncovers a novel class of lanthipeptides.
Kloosterman AM, Cimermancic P, Elsayed SS, Du C, Hadjithomas M, Donia MS, Fischbach MA, van Wezel GP, Medema MH. Kloosterman AM, et al. PLoS Biol. 2020 Dec 22;18(12):e3001026. doi: 10.1371/journal.pbio.3001026. eCollection 2020 Dec. PLoS Biol. 2020. PMID: 33351797 Free PMC article. - [Recent advances in lanthipeptide biosynthesis - A review].
Mo T, Xue L, Zhang Q. Mo T, et al. Wei Sheng Wu Xue Bao. 2016 Mar 4;56(3):373-82. Wei Sheng Wu Xue Bao. 2016. PMID: 27382781 Review. Chinese.
Cited by
- Emulating nonribosomal peptides with ribosomal biosynthetic strategies.
Mordhorst S, Ruijne F, Vagstad AL, Kuipers OP, Piel J. Mordhorst S, et al. RSC Chem Biol. 2022 Dec 6;4(1):7-36. doi: 10.1039/d2cb00169a. eCollection 2023 Jan 4. RSC Chem Biol. 2022. PMID: 36685251 Free PMC article. Review. - Structural and spectroscopic analyses of the sporulation killing factor biosynthetic enzyme SkfB, a bacterial AdoMet radical sactisynthase.
Grell TAJ, Kincannon WM, Bruender NA, Blaesi EJ, Krebs C, Bandarian V, Drennan CL. Grell TAJ, et al. J Biol Chem. 2018 Nov 9;293(45):17349-17361. doi: 10.1074/jbc.RA118.005369. Epub 2018 Sep 14. J Biol Chem. 2018. PMID: 30217813 Free PMC article. - Generation of Lasso Peptide-Based ClpP Binders.
Malik IT, Hegemann JD, Brötz-Oesterhelt H. Malik IT, et al. Int J Mol Sci. 2021 Dec 31;23(1):465. doi: 10.3390/ijms23010465. Int J Mol Sci. 2021. PMID: 35008890 Free PMC article. - N-Cα Bond Cleavage Catalyzed by a Multinuclear Iron Oxygenase from a Divergent Methanobactin-like RiPP Gene Cluster.
Chioti VT, Clark KA, Ganley JG, Han EJ, Seyedsayamdost MR. Chioti VT, et al. J Am Chem Soc. 2024 Mar 20;146(11):7313-7323. doi: 10.1021/jacs.3c11740. Epub 2024 Mar 7. J Am Chem Soc. 2024. PMID: 38452252 Free PMC article. - Isolation and Analysis of the Nisin Biosynthesis Complex NisBTC: further Insights into Their Cooperative Action.
Chen J, Kuipers OP. Chen J, et al. mBio. 2021 Oct 26;12(5):e0258521. doi: 10.1128/mBio.02585-21. Epub 2021 Oct 5. mBio. 2021. PMID: 34607454 Free PMC article.
References
- Cotter PD, Ross RP, Hill C. Bacteriocins – a viable alternative to antibiotics? Nat Rev Micro. 2013;11:95–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM097142/GM/NIGMS NIH HHS/United States
- T32 GM070421/GM/NIGMS NIH HHS/United States
- 1R01 GM097142/GM/NIGMS NIH HHS/United States
- 2T32 GM070421/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases