Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index - PubMed (original) (raw)
. 2015 Oct;47(10):1114-20.
doi: 10.1038/ng.3390. Epub 2015 Aug 31.
Andrew Bakshi 1, Zhihong Zhu 1, Gibran Hemani 1 3, Anna A E Vinkhuyzen 1, Sang Hong Lee 1 4, Matthew R Robinson 1, John R B Perry 5, Ilja M Nolte 6, Jana V van Vliet-Ostaptchouk 6 7, Harold Snieder 6; LifeLines Cohort Study; Tonu Esko 8 9 10 11, Lili Milani 8, Reedik Mägi 8, Andres Metspalu 8 12, Anders Hamsten 13, Patrik K E Magnusson 14, Nancy L Pedersen 14, Erik Ingelsson 15 16, Nicole Soranzo 17 18, Matthew C Keller 19 20, Naomi R Wray 1, Michael E Goddard 21 22, Peter M Visscher 1 2
Affiliations
- PMID: 26323059
- PMCID: PMC4589513
- DOI: 10.1038/ng.3390
Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index
Jian Yang et al. Nat Genet. 2015 Oct.
Abstract
We propose a method (GREML-LDMS) to estimate heritability for human complex traits in unrelated individuals using whole-genome sequencing data. We demonstrate using simulations based on whole-genome sequencing data that ∼97% and ∼68% of variation at common and rare variants, respectively, can be captured by imputation. Using the GREML-LDMS method, we estimate from 44,126 unrelated individuals that all ∼17 million imputed variants explain 56% (standard error (s.e.) = 2.3%) of variance for height and 27% (s.e. = 2.5%) of variance for body mass index (BMI), and we find evidence that height- and BMI-associated variants have been under natural selection. Considering the imperfect tagging of imputation and potential overestimation of heritability from previous family-based studies, heritability is likely to be 60-70% for height and 30-40% for BMI. Therefore, the missing heritability is small for both traits. For further discovery of genes associated with complex traits, a study design with SNP arrays followed by imputation is more cost-effective than whole-genome sequencing at current prices.
Conflict of interest statement
Competing Financial Interests
The authors declare no competing financial interests.
Figures
Figure 1
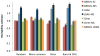
Estimates of heritability using sequence variants under different simulation scenarios based on the UK10K-WGS data. Each column represents the mean estimate from 200 simulations. Each error bar is the s.e. of the mean. The true heritability parameter is 0.8 for the simulated trait (see Online Methods for the 4 simulation scenarios).
Figure 2
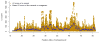
Fitting region-specific LD heterogeneity of the genome using a sliding-window approach. Shown are the results for chromosome 22 from the UK10K-WGS data as example. LD score of each variant is defined as the sum of LD _r_2 between the target variant and all variants (including the target variant) within ±10Mb distance. For the GREML-LDMS analysis, the region-specific LD heterogeneity is fitted by segments with average length of 100Kb (the dots in blue) using a sliding window approach as described in Online Methods.
Figure 3
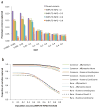
Proportion of variation at sequence variants captured by 1KGP imputation in the UK10K-WGS data. The results are the averages from 200 simulations (Online Methods). Panel (a): estimates of proportion of phenotypic variance explained by 1KGP-imputed variants in different MAF groups from GREML-MS. The 1KGP imputation was based on variants on Illumina CoreExome array extracted from the UK10K-WGS data. The column in purple represents the variance explained by the causal variants. The other four columns represent the estimates using 1KGP-imputed variants filtered at 3 levels of imputation accuracy (IMPUTE-INFO) threshold. The error bar is the s.e.m.. Without filtering variants for IMPUTE-INFO (columns in yellow), the sum of the estimate is 96.2% for common variants and 73.4% for rare variants. Panel (b): estimates of proportion of variation at sequence variants captured by 1KGP imputation (the estimate of phenotypic variance explained by the 1KGP-imputed variants summed over MAF groups divided by that explained by the causal variants) based on different types of SNP genotyping arrays. Common: MAF > 0.01; Rare: 0.01 ≥ MAF > 0.0003.
Figure 4
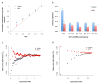
Evidence for height- and BMI-associated genetic variants being under natural selection. Results shown in panels (a) and (b) are from the GREML-LDMS analyses (Online Methods). Panel (a): the estimate of cumulative contribution of variants with MAF ≤ θ to the genetic variance, i.e. σ^v2(MAF≤θ)/σ^v2(MAF≤0.5). The dash line represents that expected under a neutral model. Panel (b): the estimate of h1KGP2 for variants in each MAF group. Error bar is s.e. of the estimate. Results shown on panel (c) are from genome-wide association analyses in the combined data set (Online Methods). bm is defined as the effect size of the minor allele of a variant. Variants are stratified into 100 MAF bins (100 quartiles of the MAF distribution). Plotted is the mean of b̂m against log10(mean MAF) in each bin. The correlation between mean b̂m and log10(mean MAF) is 0.77 (P < 1.0×10−6) for height and −0.39 (_P_ = 8.0×10−6) for BMI. Shown on panel (d) are the results from the latest GIANT consortium meta-analyses for height and BMI (see **Web Resources**) for common SNPs (MAF > 0.01). There are ~2.5M SNPs stratified into 20 MAF bins. The correlation between mean b̂m and log10(mean MAF) is 0.89 (_P_permu < 1.0×10−6) for height and −0.87 (_P_permu < 1.0×10−6) for BMI. The mean b̂m seems smaller in panel (c) than that in panel (d) because of the smaller MAF range of each bin and larger number of variants in each bin in panel (c) than those in panel (d).
Figure 5
Single-variant tagging of sequence variants by 1KGP-imputed variants. Single-variant tagging is defined as the squared correlation (_r_2max) between a sequence variant and the best tagging variant from 1KGP imputation within ±1Mb distance. Shown are the average _r_2max of variants in MAF bins for 10,000 sequence variants randomly sampled from the UK10K-WGS data. The 1KGP imputation analyses are based on variants on Illumina OmniExpress (red) and Illumina CoreExome (blue) arrays extracted from the UK10K-WGS data (see Online Methods for details about the imputation analyses based on the UK10K-WGS data). Panel (a): rare variants. Panel (b): common variants.
Similar articles
- Estimation of Genetic Relationships Between Individuals Across Cohorts and Platforms: Application to Childhood Height.
Fedko IO, Hottenga JJ, Medina-Gomez C, Pappa I, van Beijsterveldt CE, Ehli EA, Davies GE, Rivadeneira F, Tiemeier H, Swertz MA, Middeldorp CM, Bartels M, Boomsma DI. Fedko IO, et al. Behav Genet. 2015 Sep;45(5):514-28. doi: 10.1007/s10519-015-9725-7. Epub 2015 Jun 3. Behav Genet. 2015. PMID: 26036992 Free PMC article. - Genome partitioning of genetic variation for complex traits using common SNPs.
Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Yang J, et al. Nat Genet. 2011 Jun;43(6):519-25. doi: 10.1038/ng.823. Epub 2011 May 8. Nat Genet. 2011. PMID: 21552263 Free PMC article. - Ubiquitous polygenicity of human complex traits: genome-wide analysis of 49 traits in Koreans.
Yang J, Lee T, Kim J, Cho MC, Han BG, Lee JY, Lee HJ, Cho S, Kim H. Yang J, et al. PLoS Genet. 2013;9(3):e1003355. doi: 10.1371/journal.pgen.1003355. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505390 Free PMC article. - Heritability Estimation Approaches Utilizing Genome-Wide Data.
Srivastava AK, Williams SM, Zhang G. Srivastava AK, et al. Curr Protoc. 2023 Apr;3(4):e734. doi: 10.1002/cpz1.734. Curr Protoc. 2023. PMID: 37068172 Free PMC article. Review. - Recent progress in the study of the genetics of height.
Lettre G. Lettre G. Hum Genet. 2011 May;129(5):465-72. doi: 10.1007/s00439-011-0969-x. Epub 2011 Feb 22. Hum Genet. 2011. PMID: 21340692 Review.
Cited by
- Genome-wide association study reveals novel loci associated with feeding behavior in Pekin ducks.
Li GS, Zhu F, Zhang F, Yang FX, Hao JP, Hou ZC. Li GS, et al. BMC Genomics. 2021 May 8;22(1):334. doi: 10.1186/s12864-021-07668-1. BMC Genomics. 2021. PMID: 33964893 Free PMC article. - ExPRSweb: An online repository with polygenic risk scores for common health-related exposures.
Ma Y, Patil S, Zhou X, Mukherjee B, Fritsche LG. Ma Y, et al. Am J Hum Genet. 2022 Oct 6;109(10):1742-1760. doi: 10.1016/j.ajhg.2022.09.001. Epub 2022 Sep 23. Am J Hum Genet. 2022. PMID: 36152628 Free PMC article. - Genomic structural equation modelling provides a whole-system approach for the future crop breeding.
He T, Angessa TT, Hill CB, Zhang XQ, Chen K, Luo H, Wang Y, Karunarathne SD, Zhou G, Tan C, Wang P, Westcott S, Li C. He T, et al. Theor Appl Genet. 2021 Sep;134(9):2875-2889. doi: 10.1007/s00122-021-03865-4. Epub 2021 May 31. Theor Appl Genet. 2021. PMID: 34059938 - Uncovering the Genetic Architectures of Quantitative Traits.
Lee JJ, Vattikuti S, Chow CC. Lee JJ, et al. Comput Struct Biotechnol J. 2015 Nov 23;14:28-34. doi: 10.1016/j.csbj.2015.10.002. eCollection 2016. Comput Struct Biotechnol J. 2015. PMID: 27076877 Free PMC article. Review. - The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease.
Kessler T, Vilne B, Schunkert H. Kessler T, et al. EMBO Mol Med. 2016 Jul 1;8(7):688-701. doi: 10.15252/emmm.201506174. Print 2016 Jul. EMBO Mol Med. 2016. PMID: 27189168 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
- 102215/WT_/Wellcome Trust/United Kingdom
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/2/MRC_/Medical Research Council/United Kingdom
- MC_U106179472/MRC_/Medical Research Council/United Kingdom
- R01 MH100141/MH/NIMH NIH HHS/United States
- R01MH100141/MH/NIMH NIH HHS/United States
- P01 GM099568/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources