Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS - PubMed (original) (raw)
Comparative Study
Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS
Helene Tran et al. Neuron. 2015.
Abstract
Dipeptide repeat (DPR) proteins are toxic in various models of FTD/ALS with GGGGCC (G4C2) repeat expansion. However, it is unclear whether nuclear G4C2 RNA foci also induce neurotoxicity. Here, we describe a Drosophila model expressing 160 G4C2 repeats (160R) flanked by human intronic and exonic sequences. Spliced intronic 160R formed nuclear G4C2 sense RNA foci in glia and neurons about ten times more abundantly than in human neurons; however, they had little effect on global RNA processing and neuronal survival. In contrast, highly toxic 36R in the context of poly(A)(+) mRNA were exported to the cytoplasm, where DPR proteins were produced at >100-fold higher level than in 160R flies. Moreover, the modest toxicity of intronic 160R expressed at higher temperature correlated with increased DPR production, but not RNA foci. Thus, nuclear RNA foci are neutral intermediates or possibly neuroprotective through preventing G4C2 RNA export and subsequent DPR production.
Keywords: ALS; C9ORF72; DPR; Drosophila; FTD; RNA foci; Ran translation; repeats.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
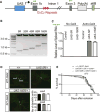
Figure 1. Expanded G4C2 Repeats in C9ORF72 Are Fully Transcribed in Patient Cells and Tissues
(A) Schematic of the three known transcript variants of wildtype (top) and mutant (bottom) alleles of C9ORF72, named as in PubMed and the UCSC Genome Browser. The locations of primers used in Panels C and D (green) and Panels E and F (blue) are indicated. (B) The G/A SNP in a control subject and in a carrier of expanded G4C2 repeats. (C, D) Pyrosequencing analysis of the allele-specific expression of V3 in three C9ORF72 and three control iPSC lines (C) and in brain tissues of three patients and three controls (D). (E, F) The relative ratio of total V1 and V3 pre-mRNAs (exon 1–intron 1 junction) to mature mRNAs (exon 1–exon 3 junction) in four C9ORF72 and four control iPSC lines (E) and in brain tissues of eight C9ORF72 patients and nine controls (F). n.s.: not statistically significant, by Student’s t test. (G) RNA-seq analysis reveals a significantly lower V2 level in patient neurons than control neurons. ***: p < 0.001, chi-square test See also Figure S1.
Figure 2. A Novel Drosophila Model of FTD/ALS Expressing Expanded G4C2 Repeats
(A) Schematic of human C9ORF72 minigenes containing 5, 80, or 160R, flanked by human intronic and exonic sequences. Green arrows: primer location used in Panel C. (B) Construction of DNA plasmids containing 5, 20, 40, 80, or 160R. Plasmids were digested with EcoRI. (C) Quantitative real-time PCR analysis of the relative levels of C9ORF72 minigene spliced RNAs. (D) Effect of 160R on dendritic branching of dda neurons in third instar larvae. No statistical difference was observed (n >14 flies per genotype). (E) Kaplan-Meier curves showing the lifespan of transgenic flies expressing 0, 5R or 160R in glutamatergic neurons by OK371-Gal4. No statistical difference between 5R and 160R flies (log-rank test). See also Figure S2.
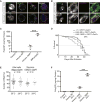
Figure 3. Overexpressed Intronic 160R Form Abundant Nuclear G4C2 Sense RNA Foci in Glia and Neurons
(A, B) The presence of multiple nuclear RNA foci (red) in glial cells (A) and glutamatergic neurons (B). The nucleus was stained with DAPI (blue) and anti-Repo (green) for glia or anti-phospho-RNA polymerase-II for glutamatergic neurons. (C) RNA FISH in muscle cells (top) and salivary gland cells (bottom) detects one single brightdot (red). (D) In each salivary gland cell expressing two copies of 160R, two brightdots are present on DNA (blue) at the site of transcription. (E) Correlation plot showing the 50 DEGs common between flies expressing 160R from the second and the third chromosome with 45 in the same direction. Two-fold change is designated by dashed red lines and genes with over two-fold change in both groups are highlighted in red. The linear regression line has a slope of 0.61, in close agreement with the relative expression levels of the transgenes in these two genetic backgrounds (Table S1). (F) Multiple RNA foci (red) in glia do not co-localize with the nucleolus-specific marker fibrillarin (green). In all panels, red arrows indicate transgene transcription site and scale bars are 5 μm. See also Figures S2F, Figure S3 and Table S1–4.
Figure 4. 36R RNAs Expressed in the Context of Poly(A)+ mRNA Are Exported to the Cytoplasm and Produce Much Higher Level of DPRs than Intronic 160R
(A) RNA FISH in glial cells detects numerous nuclear RNA foci (red) formed by intronic 160R while 36R-poly(A) RNA is exported to the cytoplasm. Green: nuclear membrane; blue: DAPI staining. Red arrows: transcription sites of transgenes. Scale bar: 5 μm. (B) Poly(GR) (red) was detected mostly in the cytoplasm of neurons expressing 36R-poly(A) but not in cells expressing intronic 160R. Scale bar: 5 μm. (C) One of the two experiments to measure poly(GP) in lysates from heads of flies raised at 25°C. Values are mean ± SEM. ***: p < 0.0001 (one-way ANOVA). On average, the poly(GP) level is 122 times higher than that in 36R flies than in 160R flies. (D) The lifespan of transgenic flies grown at 29°C expressing 0, 5R or 160R in glutamatergic neurons. Statistical difference between 5R and 160R flies was assessed with the log-rank test (p < 0.001). (E) Quantification of nuclear RNA foci in neurons and glia in flies grown at 25°C and 29°C. No statistical difference was observed (by unpaired nonparametric t test). (F) Poly(GP) levels in lysates from heads of flies raised at 25°C and 29°C. Values are mean ± SEM. ***: p <0.0001 (one-way ANOVA). See also Figure S4.
Similar articles
- Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD.
Moens TG, Mizielinska S, Niccoli T, Mitchell JS, Thoeng A, Ridler CE, Grönke S, Esser J, Heslegrave A, Zetterberg H, Partridge L, Isaacs AM. Moens TG, et al. Acta Neuropathol. 2018 Mar;135(3):445-457. doi: 10.1007/s00401-017-1798-3. Epub 2018 Jan 29. Acta Neuropathol. 2018. PMID: 29380049 Free PMC article. - Insights into C9ORF72-Related ALS/FTD from Drosophila and iPSC Models.
Yuva-Aydemir Y, Almeida S, Gao FB. Yuva-Aydemir Y, et al. Trends Neurosci. 2018 Jul;41(7):457-469. doi: 10.1016/j.tins.2018.04.002. Epub 2018 May 2. Trends Neurosci. 2018. PMID: 29729808 Free PMC article. Review. - Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death.
Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, Trotti D. Wen X, et al. Neuron. 2014 Dec 17;84(6):1213-25. doi: 10.1016/j.neuron.2014.12.010. Neuron. 2014. PMID: 25521377 Free PMC article. - Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.
Wen X, Westergard T, Pasinelli P, Trotti D. Wen X, et al. Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review. - Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders.
Boivin M, Pfister V, Gaucherot A, Ruffenach F, Negroni L, Sellier C, Charlet-Berguerand N. Boivin M, et al. EMBO J. 2020 Feb 17;39(4):e100574. doi: 10.15252/embj.2018100574. Epub 2020 Jan 13. EMBO J. 2020. PMID: 31930538 Free PMC article.
Cited by
- Identification of selective and non-selective C9ORF72 targeting in vivo active siRNAs.
Gilbert JW, Kennedy Z, Godinho BMDC, Summers A, Weiss A, Echeverria D, Bramato B, McHugh N, Cooper D, Yamada K, Hassler M, Tran H, Gao FB, Brown RH Jr, Khvorova A. Gilbert JW, et al. Mol Ther Nucleic Acids. 2024 Jul 31;35(3):102291. doi: 10.1016/j.omtn.2024.102291. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39233852 Free PMC article. - The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease.
Haeusler AR, Donnelly CJ, Rothstein JD. Haeusler AR, et al. Nat Rev Neurosci. 2016 Jun;17(6):383-95. doi: 10.1038/nrn.2016.38. Epub 2016 May 6. Nat Rev Neurosci. 2016. PMID: 27150398 Free PMC article. Review. - Molecular mechanisms underlying nucleotide repeat expansion disorders.
Malik I, Kelley CP, Wang ET, Todd PK. Malik I, et al. Nat Rev Mol Cell Biol. 2021 Sep;22(9):589-607. doi: 10.1038/s41580-021-00382-6. Epub 2021 Jun 17. Nat Rev Mol Cell Biol. 2021. PMID: 34140671 Free PMC article. Review. - C9orf72 Dipeptide Repeats Cause Selective Neurodegeneration and Cell-Autonomous Excitotoxicity in Drosophila Glutamatergic Neurons.
Xu W, Xu J. Xu W, et al. J Neurosci. 2018 Aug 29;38(35):7741-7752. doi: 10.1523/JNEUROSCI.0908-18.2018. Epub 2018 Jul 23. J Neurosci. 2018. PMID: 30037833 Free PMC article. - Functional Roles of Long Non-coding RNAs in Motor Neuron Development and Disease.
Chen KW, Chen JA. Chen KW, et al. J Biomed Sci. 2020 Feb 25;27(1):38. doi: 10.1186/s12929-020-00628-z. J Biomed Sci. 2020. PMID: 32093746 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01NS077402/NS/NINDS NIH HHS/United States
- R21 NS089979/NS/NINDS NIH HHS/United States
- R21NS089979/NS/NINDS NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- R21NS086318/NS/NINDS NIH HHS/United States
- P50AG016574/AG/NIA NIH HHS/United States
- R01NS057553/NS/NINDS NIH HHS/United States
- R01 NS063964/NS/NINDS NIH HHS/United States
- R21 NS086318/NS/NINDS NIH HHS/United States
- R01NS063964/NS/NINDS NIH HHS/United States
- R01 NS088689/NS/NINDS NIH HHS/United States
- R01NS088689/NS/NINDS NIH HHS/United States
- R01 NS057553/NS/NINDS NIH HHS/United States
- P01 NS084974/NS/NINDS NIH HHS/United States
- R01NS079725/NS/NINDS NIH HHS/United States
- R21 NS084528/NS/NINDS NIH HHS/United States
- P01NS084974/NS/NINDS NIH HHS/United States
- P01HD078253/HD/NICHD NIH HHS/United States
- R01 NS077402/NS/NINDS NIH HHS/United States
- R21NS084528/NS/NINDS NIH HHS/United States
- R01 NS079725/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous