The Replication Initiation Protein Sld3/Treslin Orchestrates the Assembly of the Replication Fork Helicase during S Phase - PubMed (original) (raw)
The Replication Initiation Protein Sld3/Treslin Orchestrates the Assembly of the Replication Fork Helicase during S Phase
Irina Bruck et al. J Biol Chem. 2015.
Retraction in
- The replication initiation protein Sld3/Treslin orchestrates the assembly of the replication fork helicase during S phase.
Bruck I, Kaplan DL. Bruck I, et al. J Biol Chem. 2017 Jun 16;292(24):10319. doi: 10.1074/jbc.A115.688424. J Biol Chem. 2017. PMID: 28623197 Free PMC article. No abstract available.
Abstract
The initiation of DNA replication is a highly regulated process in eukaryotic cells, and central to the process of initiation is the assembly and activation of the replication fork helicase. The replication fork helicase is comprised of CMG (Cdc45, Mcm2-7, and GINS) in eukaryotic cells, and the mechanism underlying assembly of the CMG during S phase was studied in this article. We identified a point mutation of Sld3 that is specifically defective for Mcm3 and Mcm5 interaction (sld3-m10), and also identified a point mutation of Sld3 that is specifically defective for single-stranded DNA (ssDNA) interaction (sld3-m9). Expression of wild-type levels of sld3-m9 resulted in a severe DNA replication defect with no recruitment of GINS to Mcm2-7, whereas expression of wild-type levels of sld3-m10 resulted in a severe replication defect with no Cdc45 recruitment to Mcm2-7. We propose a model for Sld3-mediated control of replication initiation, wherein Sld3 manages the proper assembly of the CMG during S phase. We also find that the biochemical functions identified for Sld3 are conserved in human Treslin, suggesting that Treslin orchestrates assembly of the CMG in human cells.
Keywords: DNA helicase; DNA replication; DNA-binding protein; cell division cycle 7-related protein kinase (Cdc7); cyclin-dependent kinase (CDK).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
FIGURE 1.
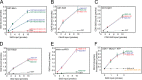
The C-terminal region of budding yeast Sld3 binds to Mcm3, Mcm5, and ssDNA. A, 10 pmol of GST-Mcm3 was used to pull down various amounts of radiolabeled Sld3, full-length and fragments, in a GST pulldown assay, as described under “Experimental Procedures.” The Sld3 had an N-terminal tag with a protein kinase A consensus sequence (LLRASV). The PKA tag was used to radiolabel the Sld3 protein with 32P for subsequent quantification. The products of the pull down were analyzed by SDS-PAGE followed by phosphorimaging, and identical experiments were averaged and plotted. B, similar to Fig. 1_A_, except GST-Mcm5 was used in place of GST-Mcm3. C, similar to Fig. 1_A_, except 10 pmol of biotinylated-ssARS1, an 80-mer ssDNA sequence from an early origin of replication, was used to pull down Sld3 in a biotin pulldown assay, as described under “Experimental Procedures.”
FIGURE 2.
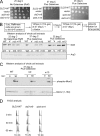
Budding yeast Sld3-m10 is defective in binding Mcm3 and Mcm5, whereas Sld3-m9 is defective in binding ssDNA. A, a GST pulldown assay similar to that described in the legend to Fig. 1, A and B. Sld3-m9 is Sld3-Q532A,F533A,S534A. Sld3-m10 is Sld3-V535A,S536A,D537A. B, similar to A. A GST pulldown assay with 10 pmol of GST-Sld3 pulling down various amounts of radiolabeled Cdc45. C, similar to A. A GST pulldown assay with 10 pmol of GST-Dpb11 pulling down various amounts of radiolabeled and CDK-phosphorylated Sld3. D, similar to A. A GST pulldown assay with 10 pmol of GST-Sld3 pulling down various amounts of radiolabeled and CDK-phosphorylated Sld2. E, similar to Fig. 1_C_. A biotin pulldown assay was performed with 10 pmol of biotin-ssARS1 pulling down various amounts of radiolabeled Sld3. F, a kinase assay was performed with 3 pmol of DDK, 3 pmol of Mcm2, [γ-32P]ATP, and various amounts of Sld3. DDK reactions were performed at 30 °C for 60 min. The products were analyzed by SDS-PAGE followed by phosphorimaging. Molecular weight markers were used to identify the position of Mcm2 in the gel. A known amount of [γ-32P]ATP was also spotted on the gel to quantify the amount of phosphate incorporation, and identical experiments were averaged and plotted. The sld3-m16 mutant was previously shown to be specifically defective in the stimulation of DDK phosphorylation of Mcm2.
FIGURE 3.
Expression of sld3-m9 and sld3-m10 at wild-type levels in budding yeast results in a severe growth defect, and markedly slowed progression through S phase. A, 10-fold serial dilution analysis of budding yeast sld3–7 td (sld3-temperature-sensitive degron) cells expressing SLD3-WT, vector, sld3-m9, or sld3-m10 from the GAL-S plasmid inducible promoter system (pRS415). The growth conditions are described at the top, and the plasmid insert is described at the left. B, Western analysis of whole cell extracts from cells used in A, probing with antibody directed against Sld3 or Arp3 (loading control). C, similar to B, except probing with antibody directed against DDK-phosphorylated Mcm2 (top gel) or Mcm2 (bottom gel). D, FACS analysis of cells described in A (37 °C plus galactose), using propidium iodide as a stain for DNA content.
FIGURE 4.
Expression of wild-type levels of sld3-m9 in budding yeast results in substantially diminished GINS-Mcm2–7 interaction during S phase, and expression of wild-type levels of sld3-m10 results in substantially diminished Cdc45-Mcm2–7 interaction during S phase. A, cells were fixed and analyzed for interaction between Sld3 and Cdc45 or Sld3 and Dpb11. Cells were synchronized in G1 with α-factor and released into medium lacking α-factor for the indicated times. Hydroxyurea was not used in these experiments. B, cells were not fixed and analyzed for interaction between Mcm2 and Sld3, Cdc45, or Psf2 (a subunit of GINS). Hydroxyurea was not used in these experiments.
FIGURE 5.
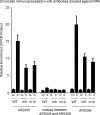
Expression of sld3-m9 and sld3-m10 at wild-type levels in budding yeast results in a slightly reduced RPA-ChIP signal at early origins of replication. Chromatin immunoprecipitation was performed using cells as described in the legend to Fig. 4_A_ (middle panel) at the restrictive temperature, in the presence of galactose. Cells were arrested with α-factor (G1 cells) and then released for 20 min (S phase cells). Cells extracts were fixed and immunoprecipitated with antibodies directed against RPA. No hydroxyurea was used in these experiments. The immunoprecipitate was probed for DNA sequence using quantitative PCR at two early origins (ARS305 or ARS306) and at a region midway between these origins. Results from repeated experiments were quantified and plotted.
FIGURE 6.
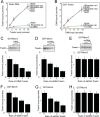
Human GINS or single-stranded DNA disrupts the interaction between Treslin and Mcm3 or Mcm5. A, similar to Fig. 1_C_. 10 pmol of biotin-DNA was used to pulldown various amounts of radiolabeled Treslin, as described under “Experimental Procedures.” Treslin had an N-terminal tag with a protein kinase A consensus sequence (LLRASV). The PKA tag was used to radiolabel the Treslin protein with 32P for subsequent quantification. The products of the pulldown were analyzed by SDS-PAGE followed by phosphorimaging, and identical experiments were averaged and plotted. B, similar to Fig. 1_A_. 10 pmol of GST-Treslin was used to pulldown various amounts of radiolabeled DNA. The sequences of the random DNA are described under “Experimental Procedures.” C, 3 pmol of GST-Mcm3 was used to pull down 3 pmol of radiolabeled Treslin in the presence of various amounts of unlabeled GINS protein. The products of the pull down were analyzed by SDS-PAGE followed by phosphorimaging, and identical experiments were averaged and plotted. D, similar to C, except random 80-mer ssDNA (strand 1) was used instead of GINS. E, similar to C, except random 80-mer dsDNA was used instead of GINS. F, similar to C, except GST-Mcm5 was used instead of GST-Mcm3. G, similar to D, except GST-Mcm5 was used instead of GST-Mcm3. H, similar to E, except GST-Mcm5 was used instead of GST-Mcm3.
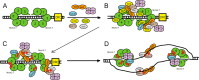
FIGURE 7.
Sld3 orchestrates the assembly of the replication fork helicase in budding yeast during S phase. A, the Mcm2–7 complex is loaded as a double hexamer in G1 phase by ORC, Cdc6 (not shown), and Cdt1 (not shown). B, two subunits of Sld3 form a tetramer with two subunits of Sld7 (not shown). DDK is recruited to Mcm2–7 during S phase. CDK is recruited to Sld2 and Sld3 during S phase. Sld3 recruits Cdc45 to Mcm2–7, and Sld3 also stimulates DDK phosphorylation of Mcm2. C, DDK phosphorylation of Mcm2–7 opens the Mcm2-Mcm5 gate, allowing for the extrusion of ssDNA from the central channel of Mcm2–7. D, once ssDNA is extruded from the central channel of Mcm2–7, the Sld3-Sld2-Dpb11 complex releases from Mcm2–7, and binds to ssDNA instead. This allows for the attachment of GINS to Mcm2–7 by a passive, sequestration mechanism. The interaction between Sld3 and ssDNA is required for the association of GINS with Mcm2–7. The helicase is now fully assembled.
Similar articles
- Insights into the Initiation of Eukaryotic DNA Replication.
Bruck I, Perez-Arnaiz P, Colbert MK, Kaplan DL. Bruck I, et al. Nucleus. 2015;6(6):449-54. doi: 10.1080/19491034.2015.1115938. Epub 2015 Dec 28. Nucleus. 2015. PMID: 26710261 Free PMC article. - Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase.
Bruck I, Kaplan DL. Bruck I, et al. Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11223-8. doi: 10.1073/pnas.1509608112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305950 Free PMC article. - Dpb11 protein helps control assembly of the Cdc45·Mcm2-7·GINS replication fork helicase.
Dhingra N, Bruck I, Smith S, Ning B, Kaplan DL. Dhingra N, et al. J Biol Chem. 2015 Mar 20;290(12):7586-601. doi: 10.1074/jbc.M115.640383. Epub 2015 Feb 6. J Biol Chem. 2015. PMID: 25659432 Free PMC article. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review. - Helicase activation and establishment of replication forks at chromosomal origins of replication.
Tanaka S, Araki H. Tanaka S, et al. Cold Spring Harb Perspect Biol. 2013 Dec 1;5(12):a010371. doi: 10.1101/cshperspect.a010371. Cold Spring Harb Perspect Biol. 2013. PMID: 23881938 Free PMC article. Review.
Cited by
- Origin DNA Melting-An Essential Process with Divergent Mechanisms.
Martinez MP, Jones JM, Bruck I, Kaplan DL. Martinez MP, et al. Genes (Basel). 2017 Jan 11;8(1):26. doi: 10.3390/genes8010026. Genes (Basel). 2017. PMID: 28085061 Free PMC article. Review. - Sld3-MCM Interaction Facilitated by Dbf4-Dependent Kinase Defines an Essential Step in Eukaryotic DNA Replication Initiation.
Fang D, Cao Q, Lou H. Fang D, et al. Front Microbiol. 2016 Jun 10;7:885. doi: 10.3389/fmicb.2016.00885. eCollection 2016. Front Microbiol. 2016. PMID: 27375603 Free PMC article. - Ensa controls S-phase length by modulating Treslin levels.
Charrasse S, Gharbi-Ayachi A, Burgess A, Vera J, Hached K, Raynaud P, Schwob E, Lorca T, Castro A. Charrasse S, et al. Nat Commun. 2017 Aug 8;8(1):206. doi: 10.1038/s41467-017-00339-4. Nat Commun. 2017. PMID: 28785014 Free PMC article. - Dpb11 may function with RPA and DNA to initiate DNA replication.
Bruck I, Dhingra N, Martinez MP, Kaplan DL. Bruck I, et al. PLoS One. 2017 May 3;12(5):e0177147. doi: 10.1371/journal.pone.0177147. eCollection 2017. PLoS One. 2017. PMID: 28467467 Free PMC article. - Cell-specific network analysis of human folliculogenesis reveals network rewiring in antral stage oocytes.
Wang S, Gong Y, Wang Z, Greenbaum J, Xiao HM, Deng HW. Wang S, et al. J Cell Mol Med. 2021 Mar;25(6):2851-2860. doi: 10.1111/jcmm.16315. Epub 2021 Feb 18. J Cell Mol Med. 2021. PMID: 33599396 Free PMC article.
References
- Tognetti S., Riera A., and Speck C. (2015) Switch on the engine: how the eukaryotic replicative helicase MCM2–7 becomes activated. Chromosoma 124, 13–26 - PubMed
- Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., and Oda M. (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous