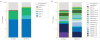An updated evolutionary classification of CRISPR-Cas systems - PubMed (original) (raw)
Review
. 2015 Nov;13(11):722-36.
doi: 10.1038/nrmicro3569. Epub 2015 Sep 28.
Yuri I Wolf 1, Omer S Alkhnbashi 2, Fabrizio Costa 2, Shiraz A Shah 3, Sita J Saunders 2, Rodolphe Barrangou 4, Stan J J Brouns 5, Emmanuelle Charpentier 6, Daniel H Haft 1, Philippe Horvath 7, Sylvain Moineau 8, Francisco J M Mojica 9, Rebecca M Terns 10, Michael P Terns 10, Malcolm F White 11, Alexander F Yakunin 12, Roger A Garrett 3, John van der Oost 5, Rolf Backofen 2 13, Eugene V Koonin 1
Affiliations
- PMID: 26411297
- PMCID: PMC5426118
- DOI: 10.1038/nrmicro3569
Review
An updated evolutionary classification of CRISPR-Cas systems
Kira S Makarova et al. Nat Rev Microbiol. 2015 Nov.
Abstract
The evolution of CRISPR-cas loci, which encode adaptive immune systems in archaea and bacteria, involves rapid changes, in particular numerous rearrangements of the locus architecture and horizontal transfer of complete loci or individual modules. These dynamics complicate straightforward phylogenetic classification, but here we present an approach combining the analysis of signature protein families and features of the architecture of cas loci that unambiguously partitions most CRISPR-cas loci into distinct classes, types and subtypes. The new classification retains the overall structure of the previous version but is expanded to now encompass two classes, five types and 16 subtypes. The relative stability of the classification suggests that the most prevalent variants of CRISPR-Cas systems are already known. However, the existence of rare, currently unclassifiable variants implies that additional types and subtypes remain to be characterized.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
Figure 1. Functional classification of Cas proteins
Protein names follow the current nomenclature and classification. An asterisk indicates that the putative small subunit (SS) protein is instead fused to Cas8 (the type I system large subunit (LS)) in several type I subtypes. The type III system LS and type IV system LS are Cas10 and Csf1 (a Cas8 family protein), respectively. Dispensable components are indicated by dashed outlines. Cas6 is shown with a solid outline for type I because it is dispensable in some but not most systems and by a dashed line for type III because most systems lack this gene and use the Cas6 provided in trans by other CRISPR–cas loci. The two colours for Cas4 and three colours for Cas9 reflect that these proteins contribute to different stages of the CRISPR Cas response. The functions shown for type IV and type V system components are proposed based on homology to the cognate components of other systems, and have not yet been experimentally verified. The functional assignments for Cpf1 are tentatively inferred by analogy with Cas9 (only the RuvC (and TnpB)-like domains of the two proteins are homologous). CARF, CRISPR-associated Rossmann fold; pre-crRNA, pre-CRISPR RNA. This research was originally published in Biochem. Soc. Trans. Makarova K. S., Wolf Y. I., & Koonin E. V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013; 41: 1392–1400 © The Biochemical Society.
Figure 2. Architectures of the genomic loci for the subtypes of CRISPR–Cas systems
Typical operon organization is shown for each CRISPR–Cas system subtype. For each representative genome, the respective gene locus tag names are indicated for each subunit. Homologous genes are colour-coded and identified by a family name. The gene names follow the classification from REF. . Where both a systematic name and a legacy name are commonly used, the legacy name is given under the systematic name. The small subunit is encoded by either csm2, cmr5, cse2 or csa5; no all-encompassing name has been proposed to collectively describe this gene family to date. Crosses through genes encoding the large subunit (Cas8 or Cas10 family members) indicate inactivation of the respective catalytic sites. Genes and gene regions encoding components of the interference module (CRISPR RNA (crRNA)–effector complexes or Cas9 proteins) are highlighted with a beige background. The adaptation module (cas1 and cas2) and cas6 are dispensable in subtypes III-A and III-B; in particular, they are rarely present in subtype III-B (dashed lines). Dark green denotes the CARF domain. Gene regions coloured cream represent the HD nuclease domain; the HD domain in Cas10 is distinct from that of Cas3 and Cas3″. Also coloured are the regions of cas9 that roughly correspond to the RuvC-like nuclease (lime green), HNH nuclease (yellow), recognition lobe (purple) and protospacer adjacent motif (PAM)-interacting domains (pink). The regions of cpf1 aside from the RuvC-like domain are functionally uncharacterized and are shown in grey, as is the functionally uncharacterized all1473 gene in subtype III-D.
Figure 3. Distribution of CRISPR–Cas systems in sequenced archaeal and bacterial genomes
a | Distribution by types. Chart showing the proportions of identified CRISPR–cas loci in bacterial or archaeal genomes that encode type I, type II, type III, type IV or type V CRISPR Cas systems. The proportion of loci that encode incomplete systems or that we could not classify unambiguously is also shown. b | Distribution by subtypes. Chart showing the proportions of identified CRISPR–cas loci in bacterial or archaeal genomes that encode each of the subtypes of CRISPR–Cas systems included in the new classification described in this article. Note that type IV and V loci each encompass a single subtype. The proportion of loci that encode incomplete systems or that we could not classify unambiguously is also shown.
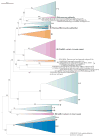
Figure 4. Comparison of different classifications of CRISPR–Cas systems
This graph shows the strength of correlation between the new classification of CRISPR–Cas systems described here (‘subtypes’; in the centre of the graph) and other classification measures. ‘Interference genes tree’ represents a phylogeny of interference module genes, which encode multisubunit CRISPR RNA (crRNA)–effector complexes or Cas9 proteins. This tree was created using a simple clustering approach based on aggregate protein sequence similarity. ‘Adaptation genes tree’ represents clustering produced by the same method but based on both components of the adaptation module, Cas1 and Cas2. ‘Cas1 phylogeny’ is the phylogenetic tree of Cas1 proteins shown in FIG. 5. ‘Loci architecture tree’ represents clustering based on a quantitative measure we developed to compare the architectures of CRISPR–cas loci. The measure is based on a weighted similarity index of the order of cas genes. ‘Repeats (sequence)’ denotes the classification of CRISPR sequences into 24 families on the basis of sequence similarity. ‘Repeats (structure)’ denotes the classification of CRISPR sequences into 18 families on the basis of structural similarity. The species tree represents the phylogeny of bacterial and archaeal translation systems. The distances depicted are inversely proportional to the degree of similarity. The full similarity matrix is shown in Supplementary information S11 (table).
Figure 5. Mapping of the CRISPR–Cas classification onto the phylogenetic tree of Cas1
Subtypes from the new classification of CRISPR–Cas systems described here were mapped onto a sequence-based phylogenetic reconstruction of 1,418 proteins from the Cas1 family, which is the most conserved Cas protein family. The phylogeny shows a close agreement with the subtype classification, as subtypes I-A, I-C, I-E, I-F, I-U, II-A, II-B, and putative type V are mostly or strictly monophyletic and are shown in gradients of light grey, except for II-B, which is shown in dark grey to indicate its origin from within I-A. The more discordant distribution of Cas1 for other subtypes probably results from horizontal transfer. None of the type III subtypes is monophyletic (in contrast to the Cas10 tree shown in Supplementary information S9 (box)), and so type III subtypes are not indicated. Note that Cas1 is absent in type IV loci and so these putative CRISPR Cas systems are not shown. Triangles denote multiple collapsed branches. Individual genes are labelled with species names and gene identification numbers. Bootstrap values are indicated as percentage points; values below 50% are not shown.
Similar articles
- Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants.
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV. Makarova KS, et al. Nat Rev Microbiol. 2020 Feb;18(2):67-83. doi: 10.1038/s41579-019-0299-x. Epub 2019 Dec 19. Nat Rev Microbiol. 2020. PMID: 31857715 Free PMC article. Review. - Diversity and evolution of class 2 CRISPR-Cas systems.
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Shmakov S, et al. Nat Rev Microbiol. 2017 Mar;15(3):169-182. doi: 10.1038/nrmicro.2016.184. Epub 2017 Jan 23. Nat Rev Microbiol. 2017. PMID: 28111461 Free PMC article. - Phylogenomics of Cas4 family nucleases.
Hudaiberdiev S, Shmakov S, Wolf YI, Terns MP, Makarova KS, Koonin EV. Hudaiberdiev S, et al. BMC Evol Biol. 2017 Nov 28;17(1):232. doi: 10.1186/s12862-017-1081-1. BMC Evol Biol. 2017. PMID: 29179671 Free PMC article. - Diversity, classification and evolution of CRISPR-Cas systems.
Koonin EV, Makarova KS, Zhang F. Koonin EV, et al. Curr Opin Microbiol. 2017 Jun;37:67-78. doi: 10.1016/j.mib.2017.05.008. Epub 2017 Jun 9. Curr Opin Microbiol. 2017. PMID: 28605718 Free PMC article. Review. - Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis.
Shmakov SA, Makarova KS, Wolf YI, Severinov KV, Koonin EV. Shmakov SA, et al. Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):E5307-E5316. doi: 10.1073/pnas.1803440115. Epub 2018 May 21. Proc Natl Acad Sci U S A. 2018. PMID: 29784811 Free PMC article.
Cited by
- Gene-Editing Technologies Paired With Viral Vectors for Translational Research Into Neurodegenerative Diseases.
Rittiner JE, Moncalvo M, Chiba-Falek O, Kantor B. Rittiner JE, et al. Front Mol Neurosci. 2020 Aug 12;13:148. doi: 10.3389/fnmol.2020.00148. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32903507 Free PMC article. - Exaptive origins of regulated mRNA decay in eukaryotes.
Hamid FM, Makeyev EV. Hamid FM, et al. Bioessays. 2016 Sep;38(9):830-8. doi: 10.1002/bies.201600100. Epub 2016 Jul 20. Bioessays. 2016. PMID: 27438915 Free PMC article. Review. - Fighting viruses with antibiotics: an overlooked path.
Colson P, Raoult D. Colson P, et al. Int J Antimicrob Agents. 2016 Oct;48(4):349-52. doi: 10.1016/j.ijantimicag.2016.07.004. Epub 2016 Aug 5. Int J Antimicrob Agents. 2016. PMID: 27546219 Free PMC article. No abstract available. - A review on molecular scissoring with CRISPR/Cas9 genome editing technology.
Irfan M, Majeed H, Iftikhar T, Ravi PK. Irfan M, et al. Toxicol Res (Camb). 2024 Jul 12;13(4):tfae105. doi: 10.1093/toxres/tfae105. eCollection 2024 Aug. Toxicol Res (Camb). 2024. PMID: 39006883 Review. - CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases.
Jolany Vangah S, Katalani C, Booneh HA, Hajizade A, Sijercic A, Ahmadian G. Jolany Vangah S, et al. Biol Proced Online. 2020 Sep 14;22:22. doi: 10.1186/s12575-020-00135-3. eCollection 2020. Biol Proced Online. 2020. PMID: 32939188 Free PMC article. Review.
References
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM99876/GM/NIGMS NIH HHS/United States
- R01 GM054682/GM/NIGMS NIH HHS/United States
- R35 GM118160/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 GM54682/GM/NIGMS NIH HHS/United States
- R01 GM099876/GM/NIGMS NIH HHS/United States
- BB/M000400/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases