Pur-alpha functionally interacts with FUS carrying ALS-associated mutations - PubMed (original) (raw)
. 2015 Oct 22;6(10):e1943.
doi: 10.1038/cddis.2015.295.
V Piccinni 1 2, V Gerbino 3, F Mantoni 1 2, S Camerini 4, J Lenzi 5, A Rosa 2, L Chellini 6, F Loreni 6, M T Carrì 3 6, I Bozzoni 2 5, M Cozzolino 3 7, G Cestra 1 2
Affiliations
- PMID: 26492376
- PMCID: PMC4632316
- DOI: 10.1038/cddis.2015.295
Pur-alpha functionally interacts with FUS carrying ALS-associated mutations
M Di Salvio et al. Cell Death Dis. 2015.
Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder due to motor neuron loss. Fused in sarcoma (FUS) protein carrying ALS-associated mutations localizes to stress granules and causes their coalescence into larger aggregates. Here we show that Pur-alpha physically interacts with mutated FUS in an RNA-dependent manner. Pur-alpha colocalizes with FUS carrying mutations in stress granules of motoneuronal cells differentiated from induced pluripotent stem cells and that are derived from ALS patients. We observe that both Pur-alpha and mutated FUS upregulate phosphorylation of the translation initiation factor eukaryotic translation initiation factor 2 alpha and consistently inhibit global protein synthesis. In vivo expression of Pur-alpha in different Drosophila tissues significatively exacerbates the neurodegeneration caused by mutated FUS. Conversely, the downregulation of Pur-alpha in neurons expressing mutated FUS significatively improves fly climbing activity. All these findings suggest that Pur-alpha, through the control of mRNA translation, might be involved in the pathogenesis of ALS associated with the mutation of FUS, and that an alteration of protein synthesis may be directly implicated in the disease. Finally, in vivo RNAi-mediated ablation of Pur-alpha produced locomotion defects in Drosophila, indicating a pivotal role for this protein in the motoneuronal function.
Figures
Figure 1
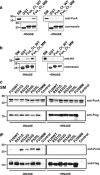
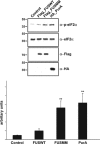
FUS/Pur-alpha physical interaction. (a) GST, GST-fused C-terminal region of wild-type FUS (FUS_Ct_WT), and GST C-terminal domain of multimutated FUS (FUS_Ct_MM) were used as baits in affinity purification experiments from a rat brain Triton X-100 extract, in the presence or absence of RNAse. Affinity-purified material retained by the GST fusion proteins was resolved by SDS-PAGE and processed by western blotting with anti-Pur-alpha antibody (top). The same volume of eluted material analyzed by western blotting was separated on a different SDS-PAGE and stained with Coomassie blue to verify equal loading of the different GST fusion baits (bottom). 1 : 500 of total brain extract and 1:50 of proteins retained from each column were loaded on the gel. SM, starting material. (b) Interaction between the same GST fusion proteins utilized in (a) and _in vitro_-translated HA-tagged Pur-alpha was tested by pull-down in the presence or absence of RNAse. HA Pur-alpha bound to the GST fusion proteins was resolved by SDS-PAGE and analyzed by western blotting with anti-HA antibody. GST fusion proteins used in the pull-down assay were resolved by SDS-PAGE and stained with Coomassie blue (bottom). 1 : 50 of reticulocyte extract exploited in the pull down and 1:3 of proteins retained from each column were loaded on the gel. (c) Protein extracts from HeLa cells expressing HA-Pur-alpha on its own (Control), or together with FUSWT, FUS carrying single mutations (R521G, R522G, R524S, or P525L), or FUSMM were incubated with or without RNAse and immunoprecipitated with anti-Flag antibody. Retained proteins were separated by SDS-PAGE and analyzed by western blotting with anti-HA and anti-Flag antibodies. 1 : 50 of total cell extract utilized for each immunoprecipitation and 1 : 3 of bound proteins were loaded on the gel. SM, starting material; IP, immunoprecipitate
Figure 2
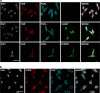
Immunofluorescence staining of FUS and Pur-alpha proteins. (a) Effect of FUSMM expression on endogenous Pur-alpha localization. Untransfected HeLa cells or cells expressing either Flag-FUSWT or Flag-FUSMM were labeled by immunofluorescence with anti-Pur-alpha, anti-Flag, and with antibodies directed against the stress granule marker TIAR (bars=10 _μ_m). (b) Untransfected HeLa cells were treated for 30 min with 1 mM of sodium arsenite and stained by immunofluorescence with anti-Pur-alpha and anti-TIAR antibodies (bars=20 _μ_m)
Figure 3
Co-localization of endogenous Pur-alpha with FUS proteins carrying single ALS-associated mutations. (a) HeLa cells transfected with plasmids encoding Flag-FUSWT or FUS carrying the indicated single C-terminal mutations were stained by immunofluorescence with anti-Pur-alpha, anti-Flag, and anti-TIAR antibodies (bars=10 _μ_m). (b) Localization of endogenous FUS and Pur-alpha in mouse spinal cord as demonstrated by the counterstain with antibodies directed against the neuronal marker NeuN (bars=20 _μ_m)
Figure 4
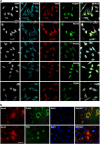
Co-localization of Pur-alpha with mutated FUS in motoneurons differentiated from IPSCs. (a) Untreated motoneurons differentiated from IPSC cells, which derived from ALS patients or from healthy donor (WT), were labeled by immunofluorescence with anti-Pur-alpha and anti-FUS antibodies (bars=10 _μ_m). (b) Same motoneurons shown in (a) were treated with 0.5 mM of sodium arsenite for 90 min to induce the formation of stress granules and stained by immunofluorescence with anti-Pur-alpha and anti-FUS antibodies (bars=10 _μ_m)
Figure 5
Effect of Pur-alpha and FUSMM expression on eIF2-alpha phosphorylation of HEK293-starved cells were transfected with HA-Pur-alpha, Flag-FUSWT, or Flag-FUSMM. eIF2-alpha phosphorylation was evaluated by western blotting with anti-p-eIF2-alpha antibody. In starved HEK293 cells the expression of HA-Pur-alpha determines a strong induction of eIF2-alpha phosphorylation as well as Flag-FUSMM, if compared with cells expressing Flag-FUSWT or untransfected cells. The lower panel shows band quantification generated with ImageJ analysis package. Data are presented as the ratio of phospho-eIF2-alpha to total eIF2-alpha signal and are expressed in arbitrary units (AU). Values are reported as mean±S.D.; _n_=3. Statistical significance was evaluated with Student's _t_-test (** indicate values significantly different from controls with _P_-value <0.001)
Figure 6
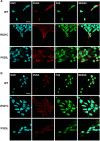
Effect of Pur-alpha and FUSMM expression on protein synthesis. (a) Untransfected HeLa cells or cells transfected with HA-Pur-alpha, Flag-FUSWT, or Flag-FUSMM were treated with puromycin. Untransfected cells are shown in the first row, while cells transfected with HA-Pur-alpha are in the second row. Cells with higher expression of Pur-alpha (transfected cells) show no incorporation of puromycin, compared with untransfected cells of the same field. Puromycin incorporation was detected by immunofluorescence with anti-puromycin antibody. Cells were also labeled by immunofluorescence with anti-Pur-alpha and anti-Flag antibodies (bars=10 _μ_m). (b) Cytoplasmic extract from HEK293 cells was separated by ultracentrifugation on a linear sucrose gradient. Eleven fractions were collected monitoring the absorbance at 260 nm and the pellet containing polyribosomes was pooled to the first fraction. The upper panel shows the absorbance profile with the position of ribosomal subunits and monomer indicated. Proteins purified from the fractions were analyzed by western blot with the indicated antibodies
Figure 7
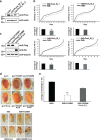
In vivo role of Pur-alpha in Drosophila melanogaster. (a) RNAi mediated downregulation of Pur-alpha in all Drosophila tissues by the expression of Pur_alpha_RNAi_1 and Pur_alpha_RNAi_2 RNAi under control of the ubiquitous driver tubulin-GAL4. Total extracts from RNAi expressing flies and control animals were separated by SDS-PAGE and the extent of Pur-alpha downregulation was evaluated by western blotting with an anti-Pur-alpha antibody. (b) The same RNAi fly lines of (a) were crossed at 29 °C with the pan neuronal driver 69B (upper panels) and with the motoneuron driver D42 (lower panels). Climbing performance of each offspring is represented by plotting the total number of climbing events for each fly of the group. Numbers of climbing events for all flies of the group were ascending ordered and plotted. Statistical significance was evaluated with Student's _t_-test (** high statistical significance, _P_-value <0.001; * statistical significance, _P_-value <0.05) and the averages of climbing events in each population, with corresponding standard errors, are shown. (c) Expression level of FUSWT, FUSMM, and Pur-alpha mammalian proteins in fly eyes. Heads from flies expressing the transgenes under GMR were separated and homogenized. Protein extracts were separated on SDS-PAGE and the expression of each transgene was evaluated by western blotting. (d) Genetic interaction of FUSMM and Pur-alpha in Drosophila eye. Eyes of flies expressing the mammalian genes under control of GMR Gal4 are shown. FUSWT and Pur-alpha induces respectively very mild and mild eye degeneration, while expressing FUSMM does not determine any visible phenotype. A simultaneous expression of both FUSMM and Pur-alpha causes strong eye degeneration. Pictures of flies expressing the transgenes under the pan neuronal driver 69B are shown. FUSWT expression causes early fly lethality (not shown), while FUSMM induces an alteration of wing extension; similar unextended wings are observed in flies expressing Pur-alpha. Expression of both FUSMM and Pur-alpha generates a more severe alteration of wing morphology. (e) Fly lines expressing FUSMM on its own, or combined with Pur_alpha_RNAi_1, were crossed with 69B-GAL4 pan neuronal driver, at 25 °C. A negative geotaxis assay was performed to measure climbing activity of flies with different genotypes. Averages of climbing evens are shown together with corresponding standard errors. Downregulation of Pur-alpha in neurons expressing FUSMM significatively improves fly climbing activity. Statistical significance was evaluated with Student's _t_-test (values significantly different from relative controls are indicated with two asterisk; P<0.001)
Similar articles
- Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity.
Daigle JG, Krishnamurthy K, Ramesh N, Casci I, Monaghan J, McAvoy K, Godfrey EW, Daniel DC, Johnson EM, Monahan Z, Shewmaker F, Pasinelli P, Pandey UB. Daigle JG, et al. Acta Neuropathol. 2016 Apr;131(4):605-20. doi: 10.1007/s00401-015-1530-0. Epub 2016 Jan 4. Acta Neuropathol. 2016. PMID: 26728149 Free PMC article. - Motor neuron apoptosis and neuromuscular junction perturbation are prominent features in a Drosophila model of Fus-mediated ALS.
Xia R, Liu Y, Yang L, Gal J, Zhu H, Jia J. Xia R, et al. Mol Neurodegener. 2012 Mar 24;7:10. doi: 10.1186/1750-1326-7-10. Mol Neurodegener. 2012. PMID: 22443542 Free PMC article. - The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span.
Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. Wang JW, et al. J Clin Invest. 2011 Oct;121(10):4118-26. doi: 10.1172/JCI57883. Epub 2011 Sep 1. J Clin Invest. 2011. PMID: 21881207 Free PMC article. - TDP-43 and FUS/TLS: sending a complex message about messenger RNA in amyotrophic lateral sclerosis?
Strong MJ, Volkening K. Strong MJ, et al. FEBS J. 2011 Oct;278(19):3569-77. doi: 10.1111/j.1742-4658.2011.08277.x. Epub 2011 Sep 6. FEBS J. 2011. PMID: 21810174 Review. - Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma.
Bentmann E, Haass C, Dormann D. Bentmann E, et al. FEBS J. 2013 Sep;280(18):4348-70. doi: 10.1111/febs.12287. Epub 2013 May 9. FEBS J. 2013. PMID: 23587065 Review.
Cited by
- Where and Why Modeling Amyotrophic Lateral Sclerosis.
Liguori F, Amadio S, Volonté C. Liguori F, et al. Int J Mol Sci. 2021 Apr 12;22(8):3977. doi: 10.3390/ijms22083977. Int J Mol Sci. 2021. PMID: 33921446 Free PMC article. Review. - RNA Dysregulation in Amyotrophic Lateral Sclerosis.
Butti Z, Patten SA. Butti Z, et al. Front Genet. 2019 Jan 22;9:712. doi: 10.3389/fgene.2018.00712. eCollection 2018. Front Genet. 2019. PMID: 30723494 Free PMC article. Review. - HDAC1 inhibition ameliorates TDP-43-induced cell death in vitro and in vivo.
Sanna S, Esposito S, Masala A, Sini P, Nieddu G, Galioto M, Fais M, Iaccarino C, Cestra G, Crosio C. Sanna S, et al. Cell Death Dis. 2020 May 14;11(5):369. doi: 10.1038/s41419-020-2580-3. Cell Death Dis. 2020. PMID: 32409664 Free PMC article. - Nuclear poly(A) binding protein 1 (PABPN1) and Matrin3 interact in muscle cells and regulate RNA processing.
Banerjee A, Vest KE, Pavlath GK, Corbett AH. Banerjee A, et al. Nucleic Acids Res. 2017 Oct 13;45(18):10706-10725. doi: 10.1093/nar/gkx786. Nucleic Acids Res. 2017. PMID: 28977530 Free PMC article. - Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells.
Ichiyanagi N, Fujimori K, Yano M, Ishihara-Fujisaki C, Sone T, Akiyama T, Okada Y, Akamatsu W, Matsumoto T, Ishikawa M, Nishimoto Y, Ishihara Y, Sakuma T, Yamamoto T, Tsuiji H, Suzuki N, Warita H, Aoki M, Okano H. Ichiyanagi N, et al. Stem Cell Reports. 2016 Apr 12;6(4):496-510. doi: 10.1016/j.stemcr.2016.02.011. Epub 2016 Mar 17. Stem Cell Reports. 2016. PMID: 26997647 Free PMC article.
References
- 3Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009; 323: 1205–1208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous