Nucleolar repression facilitates initiation and maintenance of senescence - PubMed (original) (raw)
Nucleolar repression facilitates initiation and maintenance of senescence
Leixiang Yang et al. Cell Cycle. 2015.
Abstract
Tumor cells with defective apoptosis pathways often respond to chemotherapy by entering irreversible cell cycle arrest with features of senescence. However, rare cells can bypass entry to senescence, or re-enter cell cycle from a senescent state. Deficiency in senescence induction and maintenance may contribute to treatment resistance and early relapse after therapy. Senescence involves epigenetic silencing of cell cycle genes and reduced rRNA transcription. We found that senescence-inducing treatments such as DNA damage and RNA polymerase I inhibition stimulate the binding between the nucleolar protein NML (nucleomethylin) and SirT1. The NML complex promotes rDNA heterochromatin formation and represses rRNA transcription. Depletion of NML reduced the levels of H3K9Me3 and H3K27Me3 heterochromatin markers on rDNA and E2F1 target promoters in senescent cells, increased rRNA transcription, and increased the frequency of cell cycle re-entry. Depletion of the nucleolar transcription repressor factor TIP5 also promoted escape from senescence. Furthermore, tumor tissue staining showed that breast tumors without detectable nucleolar NML expression had poor survival. The results suggest that efficient regulation of nucleolar rDNA transcription facilitates the maintenance of irreversible cell cycle arrest in senescent cells. Deficiency in nucleolar transcription repression may accelerate tumor relapse after chemotherapy.
Keywords: NML; chemotherapy; heterochromatin; nucleolus; p53; rDNA; senescence.
Figures
Figure 1.
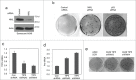
NML knockdown promotes escape from drug-induced senescence. (A) A549 cells were stably infected with retrovirus expressing NML shRNAs. NML knockdown efficiency was confirmed by protein gel blot. (B) Pre-rRNA expression levels in stable NML knockdown cell lines were determined by RT-qPCR. Values are mean ± SD of triplicates. The results are reproducible in multiple experiments. (C) A549 stable NML knockdown cells were treated with 0.1 µM doxorubicin for 7 days, cultured in drug-free medium for 7 days, and stained for SA-β-gal activity. Circled area indicates SA-β-gal-negative cells that may have bypassed senescence during drug treatment or undergone reversal after drug removal. Colonies formed over a senescent cell monolayer were stained by crystal violet at 21 d Bar=20 µm. (D) Colony formation efficiency by A549 with NML knockdown after senescence induction with 0.1 µM doxorubicin for 7 d followed by drug washout for 21 d without re-seeding. (E) A549 cells were treated with 0.1 µM CX5461 for 7 d and stained for SA-β-gal. Bar=20 µm. (F) H1299 stably infected with NML shRNA retroviruses were treated with doxorubicin for 5 d and cultured in drug-free medium for 9 d Colonies were stained with crystal violet.
Figure 2.
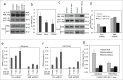
Depletion of NML in senescent cells promotes escape. (A) A549 cells were treated with 0.1 μM doxorubicin for 7 d and then transfected with NML siRNA for 2 d. NML knockdown in senescent cells was verified by western blot. (B) A549 cells were treated with 0.1 µM doxorubicin for 7 d to induce senescence, followed by transfection with NML or p53 siRNA. Colonies formed over the senescent monolayer were stained after 21 d (C) A549 cells were stably infected with retrovirus expressing TIP5 shRNA, the knockdown efficiency was confirmed by RT-qPCR. Values are mean ± SD of triplicates. (D) Pre-rRNA expression levels in A549 TIP5 knockdown cells were determined by RT-qPCR. Values are mean ± SD of 3 experiments. (E) A549 TIP5 knockdown cells were treated with 0.1 µM doxorubicin for 7 d Colony formation in drug-free medium was determined without re-seeding.
Figure 3.
NML knockdown prevents heterochromatin formation on rDNA. (A) A549 cells were treated with 0.3 µM CX5461 for 2 hrs and 24 hrs. Endogenous NML-SirT1 binding was analyzed by SirT1 IP and NML protein gel blot. (B) Pre-rRNA level in senescent cells induced by 0.1 µM doxorubicin or 0.3 µM CX5461 for 7 d was determined by RT-qPCR. Values are mean ± SD of triplicates. (C) A549 cells were incubated with indicated drugs for 4 d The endogenous SirT1-NML complex was detected by IP-western blot. (D) Pre-rRNA level in A549 NML knockdown cells treated with 0.1 µM doxorubicin for 24 and 72 hrs was analyzed by RT-qPCR. Values are mean ± SD of triplicates. (E, F) A549 NML knockdown cells were treated with 0.3 µM CX5461 for 1 and 7 d H3K9Me3 and H3K27Me3 levels on rDNA promoter were detected by ChIP and qPCR. (G) A549 expressing 2 different NML shRNA were treated with CX5461 for 7 d and analyzed for histone methylation at the rDNA promoter by ChIP.
Figure 4.
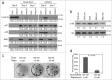
NML knockdown compromises cell cycle regulation. (A) A549 NML knockdown cells were treated with 0.1 µM doxorubicin or 0.3 µM CX5461 for 3 and 7 d. Expression levels of cell cycle markers were analyzed by protein gel blot. Numbers in boxes show pair-wise quantitation of control shRNA and NML shRNA. (B) The level of phosphorylated ribosomal protein S6 (Ser240/Ser244) was determined in A549 NML and TIP5 knockdown cells by western blot. Numbers represent relative quantitation of bands. (C) A549 cells were co-treated with doxorubicin and rapamycin for 7 d and cultured in drug-free medium for 21 d to detect colony formation over the senescent monolayer. (D) A549 cells were co-treated with doxorubicin and rapamycin for 7 d and the percentage of SA-β-gal positive cells was determined.
Figure 5.
NML depletion alters the regulation of senescence-related genes. (A) A549-control shRNA cells were treated with 0.3 µM CX5461 for 5 d and the mRNA levels of 84 senescence-related genes were analyzed by RT-PCR. Fold changes were determined by comparing treated to untreated cells. The genes were sorted from strongest induction (left) to strongest repression (right). The names of the top 18 activated genes and top 6 repressed genes are shown. (B) Comparison of senescence gene expression levels after 5 d of CX5461 treatment. The mRNA levels for control and NML knockdown cells after 5 d of CX5461 treatment were compared and the ratios were shown in the same gene order as (A). A ratio of +1 or -1 indicates the gene was induced to the same degree in control and knockdown cells. A ratio of >+1 indicates stronger induction in NML knockdown cells compared to control cells, a ratio of <-1 indicates weaker induction in NML knockdown cells compared to control cells. The results are representative of 3 experiments.
Figure 6.
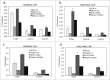
Depletion of NML reduces heterochromatin formation at E2F target genes. (A, B) A549 NML knockdown cells were treated with 0.3 µM CX5461 for 7 d. H3K9Me3 and H3K27Me3 levels at indicated E2F1 target promoters were detected by ChIP and qPCR. Values are mean ± SD of triplicates. (C, D) A549 TIP5 knockdown cells were treated with 0.3 µM CX5461 for 7 d. H3K9Me3 and H3K27Me3 levels at indicated E2F1 target promoters were detected by ChIP and qPCR. Values are mean ± SD of triplicates. All results are reproducible in multiple experiments.
Figure 7.
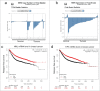
NML expression is downregulated in a subset of tumors. (A) Reduced NML gene (RRP8) copy number in a bladder cancer cohort (Oncomine). A subset of tumor samples showed heterozygous deletion at the RRP8 locus. (B) Reduced NML mRNA level in an invasive breast cancer cohort compared to normal tissue (Oncomine). (C, D) Kaplan-Meyer plots of 3450 patients with breast tumors following systemic treatment showing that low NML (RRP8, probe set 203170_at) and TIP5 (TIP5, probe set 201355_s_at) mRNA levels are associated with shorter relapse free survival.
Figure 8.
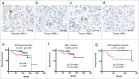
Absence of NML staining is associated with poor survival in ER-negative breast cancer. (A, B, C, D) NML expression in breast tumors was detected by IHC staining. Arrows indicate nucleolar NML staining in normal breast control and a subset of breast tumors. Bar=20 µm. (E, F, G) Kaplan-Meier plots of breast tumor survival stratified based on nucleolar NML positive and negative staining and ER status.
Similar articles
- Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription.
Holmberg Olausson K, Nistér M, Lindström MS. Holmberg Olausson K, et al. J Biol Chem. 2014 Dec 12;289(50):34601-19. doi: 10.1074/jbc.M114.569244. Epub 2014 Oct 27. J Biol Chem. 2014. PMID: 25349213 Free PMC article. - Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability.
Yang L, Song T, Chen L, Kabra N, Zheng H, Koomen J, Seto E, Chen J. Yang L, et al. Mol Cell Biol. 2013 Oct;33(19):3835-48. doi: 10.1128/MCB.00476-13. Epub 2013 Jul 29. Mol Cell Biol. 2013. PMID: 23897426 Free PMC article. - Nucleolar aggresomes mediate release of pericentric heterochromatin and nuclear destruction of genotoxically treated cancer cells.
Salmina K, Huna A, Inashkina I, Belyayev A, Krigerts J, Pastova L, Vazquez-Martin A, Erenpreisa J. Salmina K, et al. Nucleus. 2017 Mar 4;8(2):205-221. doi: 10.1080/19491034.2017.1279775. Epub 2017 Jan 9. Nucleus. 2017. PMID: 28068183 Free PMC article. - A metabolic throttle regulates the epigenetic state of rDNA.
Grummt I, Ladurner AG. Grummt I, et al. Cell. 2008 May 16;133(4):577-80. doi: 10.1016/j.cell.2008.04.026. Cell. 2008. PMID: 18485866 Review. - Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments.
Bártová E, Horáková AH, Uhlírová R, Raska I, Galiová G, Orlova D, Kozubek S. Bártová E, et al. J Histochem Cytochem. 2010 May;58(5):391-403. doi: 10.1369/jhc.2009.955435. Epub 2009 Dec 21. J Histochem Cytochem. 2010. PMID: 20026667 Free PMC article. Review.
Cited by
- Tumor cell senescence response produces aggressive variants.
Yang L, Fang J, Chen J. Yang L, et al. Cell Death Discov. 2017 Aug 21;3:17049. doi: 10.1038/cddiscovery.2017.49. eCollection 2017. Cell Death Discov. 2017. PMID: 28845296 Free PMC article. - RRP8, associated with immune infiltration, is a prospective therapeutic target in hepatocellular carcinoma.
You K, Du X, Zhao Y, Wen F, Lu Z, Fan H. You K, et al. J Cancer Res Clin Oncol. 2024 May 9;150(5):245. doi: 10.1007/s00432-024-05756-9. J Cancer Res Clin Oncol. 2024. PMID: 38722372 Free PMC article. - Cellular senescence and abdominal aortic aneurysm: From pathogenesis to therapeutics.
Wang D, Hao X, Jia L, Jing Y, Jiang B, Xin S. Wang D, et al. Front Cardiovasc Med. 2022 Sep 14;9:999465. doi: 10.3389/fcvm.2022.999465. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36187019 Free PMC article. Review.
References
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005; 120:513-22; PMID:15734683; http://dx.doi.org/10.1016/j.cell.2005.02.003 - DOI - PubMed
- d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nature Rev Cancer 2008; 8:512-22; http://dx.doi.org/10.1038/nrc2440 - DOI - PubMed
- Schmitt CA. Cellular senescence and cancer treatment. Biochimica Et Biophysica Acta 2007; 1775:5-20; PMID:17027159 - PubMed
- Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol 2011; 192:547-56; PMID:21321098; http://dx.doi.org/10.1083/jcb.201009094 - DOI - PMC - PubMed
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011; 479:232-6; PMID:22048312; http://dx.doi.org/10.1038/nature10600 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous