The architecture of a eukaryotic replisome - PubMed (original) (raw)
. 2015 Dec;22(12):976-82.
doi: 10.1038/nsmb.3113. Epub 2015 Nov 2.
Affiliations
- PMID: 26524492
- PMCID: PMC4849863
- DOI: 10.1038/nsmb.3113
The architecture of a eukaryotic replisome
Jingchuan Sun et al. Nat Struct Mol Biol. 2015 Dec.
Abstract
At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase is positioned in front to unwind DNA and that DNA polymerases trail behind the helicase. Here we used single-particle EM to directly image a Saccharomyces cerevisiae replisome. Contrary to expectations, the leading strand Pol ɛ is positioned ahead of CMG helicase, whereas Ctf4 and the lagging-strand polymerase (Pol) α-primase are behind the helicase. This unexpected architecture indicates that the leading-strand DNA travels a long distance before reaching Pol ɛ, first threading through the Mcm2-7 ring and then making a U-turn at the bottom and reaching Pol ɛ at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture and provides a basis for further structural and biochemical replisome studies.
Figures
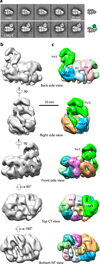
Figure 1. Structure of the S. cerevisiae CMG helicase
(a) Surface-rendered and segmented ScCMG structure in different views of the 3D EM map. The larger dashed red circle marks the apparent hole in the middle of Mcm2-7, and the smaller circle indicates the apparent second hole between GINS-Cdc45 and Mcm2-7. (b) Corresponding views of the segmented map. (c) Docking of the crystal structures of the GINS (PDB ID 2E9X), Cdc45 homolog (PDB ID 1IR6), and six copies of the Sulfolobus solfataricus MCM monomer crystal structure (PDB ID 3F9V). (d) Docking in the Mcm2-7 region of the NTD hexamer crystal structure of the Sulfolobus solfataricus MCM (PDB ID 2VL6). The green dot in (b) and (c) marks the unoccupied density between assigned GINS and Cdc45 that may be the truncated CTD of GINS subunit Psf1 (Psf1- CTD), but may also belong to Cdc45.
Figure 2. Structure of the S. cerevisiae CMGE leading strand helicase-polymerase
(a) Selected side views of reference-free class averages of CMG (top row) compared to CMG complexed with Pol ε (bottom row). (b) Surface rendered EM map in five views. Display threshold is set to include the predicted molecular mass. (c) Segmented map. The Pol ε density is shown in green. The distinct groove may be the polymerase active site and the arm of density extending down between GINS and Cdc45 is the shape expected for the Dpb3/4 histone fold heterodimer subunits. Assignments of subunits in the Pol ε density are not certain as explained in the text. The six “Zn” labels in the bottom NT view denote the N-terminal Zn finger domains of Mcm proteins (see also the Supplemental video).
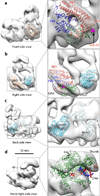
Figure 3. Rigid-body docking of CMG subunits into the CMGE density map with available crystal structures
(a) The crystal structures of human GINS complex (PDB ID 2E9X) fit well in the EM density. The four GINS subunits are colored red (Psf1), green (Psf2), blue (Psf3), and orange (Sld5) respectively. The red spheres show the last residue in the CTD-truncated Psf1 crystal structure. The magenta spheres show the N-terminal first resolved residue (Leu21) of the Sld5 subunit. (b) Side view showing the docking of human GINS and the catalytic core of the RecJ exonuclease homolog to Cdc45 (PDB ID 1IR6, cyan). (c) Back side view of the catalytic core of the RecJ exonuclease homolog to Cdc45 (PDB ID 1IR6, cyan) adjacent to the Sulfolobus solfataricus MCM monomer crystal structure (PDB ID 3F9V) labeled “M2”. (d) Crystal structure of the Pol2 catalytic N-terminal domain complexed with primed DNA and dNTP (PDB ID 4M8O) is docked such that DNA aligns into the large groove; this is speculative and should be regarded as tentative due to the underweight density of Pol ε in the CMGE structure. Template and primer strand DNA is in red and blue, respectively.
Figure 4. Subunit proximities within CMGE determined by chemical cross-linking with mass spectrometry readout (CX-MS)
CMGE was cross-linked with a lysine specific bifunctional cross-linker, then fragmented by proteolysis followed by identification of crosslinked peptides by mass spectrometry. (a) Overview of cross-links observed within the region of Pol2 corresponding to the crystal structure (PDB 4M8O). The cross-linked lysine residues are presented as red spheres. Straight lines represent DSS cross-links. (b) Intersubunit cross-links between subunits of the 15-protein CMGE complex. The lengths of the subunits correspond to the lengths of the colored rectangles that they represent: Mcms (yellow), GINS (blue), Cdc45 (purple), and Pol ε (green). (c) The upper left view illustrates major cross-links observed to connect GINS and Cdc45 to Mcm3/5 and Mcm2, respectively. Remaining views illustrate cross links between Pol ε and CMG, clockwise: Putative Dpb3/Dpb4 to Cdc45. Pol2 C-terminal region cross-links to the C-terminal regions of Mcm2 and Mcm6, and Cdc45. Dpb2 cross-links to the Mcm5 C-terminal region and to Psf1 of GINS.
Figure 5. Staged assembly of the eukaryotic replisome
(a–c). Replisome reconstitution. Selected side views of reference-free class averages of: (a) CMG mixed with Ctf4 and 80/75mer, (b) CMG mixed with Ctf4, Pol α and 80/75mer, (c) CMG mixed with Pol ε, Ctf4 and 80/75mer. (d) CMG mixed with Pol ε and the 160/91mer primed fork. (e) CMG mixed with Pol ε, Ctf4, Pol α and the 160/91mer primed fork.
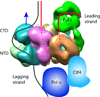
Figure 6. Architecture of the eukaryotic replisome
Replisome structure and the proposed DNA path through the replisome. Pol α is shown in blue. Ctf4 is in cyan. Red and black lines illustrate possible leading and lagging strand DNA. The blue arrow indicates the direction of replisome movement on DNA. The diagram indicates a long leading strand DNA path through the entire Mcm ring and then bending back up to Pol ε, requiring about 40 ntd of ssDNA. Leading ssDNA is illustrated as going completely through the Mcm2-7 complex, then bending up through the second “accessory” channel of CMG, but this path is speculative. Other DNA paths are possible. See text and supplementary Fig. 6 for further details.
Similar articles
- A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome.
Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinković D, Labib K, Costa A, Pellegrini L. Simon AC, et al. Nature. 2014 Jun 12;510(7504):293-297. doi: 10.1038/nature13234. Epub 2014 May 4. Nature. 2014. PMID: 24805245 Free PMC article. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review. - The eukaryotic CMG helicase pumpjack and integration into the replisome.
Sun J, Yuan Z, Georgescu R, Li H, O'Donnell M. Sun J, et al. Nucleus. 2016 Apr 25;7(2):146-54. doi: 10.1080/19491034.2016.1174800. Nucleus. 2016. PMID: 27310307 Free PMC article. Review. - Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, O'Donnell ME. Georgescu R, et al. Proc Natl Acad Sci U S A. 2017 Jan 31;114(5):E697-E706. doi: 10.1073/pnas.1620500114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096349 Free PMC article. - Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork.
Schauer GD, O'Donnell ME. Schauer GD, et al. Proc Natl Acad Sci U S A. 2017 Jan 24;114(4):675-680. doi: 10.1073/pnas.1619748114. Epub 2017 Jan 9. Proc Natl Acad Sci U S A. 2017. PMID: 28069954 Free PMC article.
Cited by
- Tissue-Specific Requirement for the GINS Complex During Zebrafish Development.
Varga M, Csályi K, Bertyák I, Menyhárd DK, Poole RJ, Cerveny KL, Kövesdi D, Barátki B, Rouse H, Vad Z, Hawkins TA, Stickney HL, Cavodeassi F, Schwarz Q, Young RM, Wilson SW. Varga M, et al. Front Cell Dev Biol. 2020 May 28;8:373. doi: 10.3389/fcell.2020.00373. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32548116 Free PMC article. - Embracing Heterogeneity: Challenging the Paradigm of Replisomes as Deterministic Machines.
Lewis JS, van Oijen AM, Spenkelink LM. Lewis JS, et al. Chem Rev. 2023 Dec 13;123(23):13419-13440. doi: 10.1021/acs.chemrev.3c00436. Epub 2023 Nov 16. Chem Rev. 2023. PMID: 37971892 Free PMC article. Review. - Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks.
Liu L, Zhang Y, Zhang J, Wang JH, Cao Q, Li Z, Campbell JL, Dong MQ, Lou H. Liu L, et al. Cell Mol Life Sci. 2020 Aug;77(15):3041-3058. doi: 10.1007/s00018-019-03333-9. Epub 2019 Nov 15. Cell Mol Life Sci. 2020. PMID: 31728581 Free PMC article. - The Role of MTBP as a Replication Origin Firing Factor.
Zaffar E, Ferreira P, Sanchez-Pulido L, Boos D. Zaffar E, et al. Biology (Basel). 2022 May 27;11(6):827. doi: 10.3390/biology11060827. Biology (Basel). 2022. PMID: 35741348 Free PMC article. Review. - Flexibility and Distributive Synthesis Regulate RNA Priming and Handoff in Human DNA Polymerase α-Primase.
Cordoba JJ, Mullins EA, Salay LE, Eichman BF, Chazin WJ. Cordoba JJ, et al. J Mol Biol. 2023 Dec 15;435(24):168330. doi: 10.1016/j.jmb.2023.168330. Epub 2023 Oct 24. J Mol Biol. 2023. PMID: 37884206 Free PMC article.
References
References for Online Methods
- Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. - PubMed
- Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG29979/AG/NIA NIH HHS/United States
- R37 GM038839/GM/NIGMS NIH HHS/United States
- GM109824/GM/NIGMS NIH HHS/United States
- R01 GM074985/GM/NIGMS NIH HHS/United States
- R01 GM111742/GM/NIGMS NIH HHS/United States
- R01 GM038839/GM/NIGMS NIH HHS/United States
- R35 GM131754/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- R01 AG029979/AG/NIA NIH HHS/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
- S10 RR025415/RR/NCRR NIH HHS/United States
- GM74985/GM/NIGMS NIH HHS/United States
- R01 GM115809/GM/NIGMS NIH HHS/United States
- GM103314/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM38839/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous