PDL1 Regulation by p53 via miR-34 - PubMed (original) (raw)
. 2015 Nov 17;108(1):djv303.
doi: 10.1093/jnci/djv303. Print 2016 Jan.
Cristina Ivan 1, David Valdecanas 1, Xiaohong Wang 1, Heidi J Peltier 1, Yuping Ye 1, Luiz Araujo 1, David P Carbone 1, Konstantin Shilo 1, Dipak K Giri 1, Kevin Kelnar 1, Desiree Martin 1, Ritsuko Komaki 1, Daniel R Gomez 1, Sunil Krishnan 1, George A Calin 1, Andreas G Bader 1, James W Welsh 2
Affiliations
- PMID: 26577528
- PMCID: PMC4862407
- DOI: 10.1093/jnci/djv303
PDL1 Regulation by p53 via miR-34
Maria Angelica Cortez et al. J Natl Cancer Inst. 2015.
Abstract
Background: Although clinical studies have shown promise for targeting PD1/PDL1 signaling in non-small cell lung cancer (NSCLC), the regulation of PDL1 expression is poorly understood. Here, we show that PDL1 is regulated by p53 via miR-34.
Methods: p53 wild-type and p53-deficient cell lines (p53(-/-) and p53(+/+) HCT116, p53-inducible H1299, and p53-knockdown H460) were used to determine if p53 regulates PDL1 via miR-34. PDL1 and miR-34a expression were analyzed in samples from patients with NSCLC and mutated p53 vs wild-type p53 tumors from The Cancer Genome Atlas for Lung Adenocarcinoma (TCGA LUAD). We confirmed that PDL1 is a direct target of miR-34 with western blotting and luciferase assays and used a p53(R172HΔ)g/+K-ras(LA1/+) syngeneic mouse model (n = 12) to deliver miR-34a-loaded liposomes (MRX34) plus radiotherapy (XRT) and assessed PDL1 expression and tumor-infiltrating lymphocytes (TILs). A two-sided t test was applied to compare the mean between different treatments.
Results: We found that p53 regulates PDL1 via miR-34, which directly binds to the PDL1 3' untranslated region in models of NSCLC (fold-change luciferase activity to control group, mean for miR-34a = 0.50, SD = 0.2, P < .001; mean for miR-34b = 0.52, SD = 0.2, P = .006; and mean for miR-34c = 0.59, SD = 0.14, and P = .006). Therapeutic delivery of MRX34, currently the subject of a phase I clinical trial, promoted TILs (mean of CD8 expression percentage of control group = 22.5%, SD = 1.9%; mean of CD8 expression percentage of MRX34 = 30.1%, SD = 3.7%, P = .016, n = 4) and reduced CD8(+)PD1(+) cells in vivo (mean of CD8/PD1 expression percentage of control group = 40.2%, SD = 6.2%; mean of CD8/PD1 expression percentage of MRX34 = 20.3%, SD = 5.1%, P = .001, n = 4). Further, MRX34 plus XRT increased CD8(+) cell numbers more than either therapy alone (mean of CD8 expression percentage of MRX34 plus XRT to control group = 44.2%, SD = 8.7%, P = .004, n = 4). Finally, miR-34a delivery reduced the numbers of radiation-induced macrophages (mean of F4-80 expression percentage of control group = 52.4%, SD = 1.7%; mean of F4-80 expression percentage of MRX34 = 40.1%, SD = 3.5%, P = .008, n = 4) and T-regulatory cells.
Conclusions: We identified a novel mechanism by which tumor immune evasion is regulated by p53/miR-34/PDL1 axis. Our results suggest that delivery of miRNAs with standard therapies, such as XRT, may represent a novel therapeutic approach for lung cancer.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
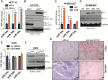
Figure 1.
p53 regulation of PDL1 via miR-34. A) miR-34a, miR-34b, and miR-34c are upregulated in HCT116 p53+/+ cells treated with nutlin 3 (10 µM for 24 hours) (P = .0001, P < .001, P < .001) and in (C) H1299 p53-inducible cells treated with ponasterone A (PoA) (5 µM for 24, 48, and 72 hours) compared with their respective controls, confirming that p53 overexpression induces miR-34a, miR-34b, and perhaps miR-34c expression (P = .001, P = .004, P = .09). An unpaired t test was used to calculate the two-sided P values. E) Downregulation of miR-34a but not miR-34b or miR-34c, in H460 cells expressing p53-targeting shRNA compared with control (P = .006, P = .19, P = .20). An unpaired t test was used to calculate the two-sided P values. B) Downregulation of PDL1 expression in HCT116 p53+/+ vs p53-/- cells and in H1299 p53-inducible cells (D) and upregulation of PDL1 in H460 cells treated with p53-shRNA (F). G) Immunohistochemical staining of samples from patients with NSCLC showing higher PDL1 protein levels (top row) in tumors with p53 mutation than in tumors with wild-type (wt) p53 (3 patients per group). Chromogenic in situ hybridization staining (CISH; bottom row) indicated downregulation of miR-34a in tumors with mutated p53 relative to tumors with wt p53. Magnification ×100. Scale bar = 100 μm. Error bars on the bar charts represent standard deviation. *P < .05, **P < .01, ***P < .001. ctrl = control; mut = mutated; PoA = ponasterone A; wt = wild-type.
Figure 2.
Correlation of p53 with PDL1 expression in patients with non–small cell lung cancer. A) Correlation between p53 and PDL1 (CD274) mRNA expression in samples from 181 patients with NSCLC from The Cancer Genome Atlas (TCGA) (P < .001) (29). The Spearman’s rank-order correlation test was applied to measure the strength of the association between p53 and PDL1 (CD274) mRNA levels. B) PDL1 expression in patients with p53-mutated tumors (n = 84) and p53 wild-type (wt) tumors (n = 97) showed that tumors with mutated p53 had higher PDL1 levels than did those with wt p53 (P = .03). CD274 levels were compared between p53 mutant tumors and p53 wt tumors with Mann-Whitney tests. C) miR-34a levels were lower in tumors with mutated p53 vs wt p53 (P = .01). A box-and-whisker plot is used to represent the data. Box plot represents first (lower bound) quartile, median and third (upper bound) quartile. Whiskers, representing 1.5 times the interquartile range, were used to visualize data (log2) for these comparisons. Mann-Whitney-Wilcoxon test. Two-sided. D) Kaplan-Meier overall survival curves according to CD274 and TP53 expression in TCGA LUAD patients cohorts. The number of patients at risk in high CD274/low TP53 and low CD274/high TP53 groups at different time points are presented at the bottom of the graph. Log-rank test, two-sided. Patients with tumors that expressed high PDL1 and low p53 levels had lower survival rates than did patients with low PDL1/high p53 tumors (P = .005). E and F) Patients with high miR-34a/high p53 or high p53-only tumors had better survival rates than did patients with low miR-34a/low p53 tumors (E) (P = .004) or low p53-only tumors (F) (P = .03). The log-rank test was used to determine the association between mRNA/miRNA expression and overall survival, and the Kaplan-Meyer method was used to generate survival curves. All tests were two-sided and considered statistically significant at the .05 level. *P < .05, **P < .01, ***P < .001. LUAD = lung adenocarcinoma; TCGA = The Cancer Genome Atlas; WT = wild-type.
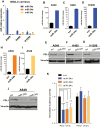
Figure 3.
miR-34s regulation of PDL1. Endogenous levels of miR-34a, -b, and -c in non–small cell lung cancer (NSCLC) cell lines. A) PDL1 expression in A549, H460, and H1299 cells transfected with miR-34a, -b, and -c. B-J) NSCLC cell lines were treated with miR-34a, miR-34b, or miR-34c at 100nM, and 24 hours later RNA was isolated to study miR-34a, -b, and -c transfection efficiency. At 96 hours after transfection, cell lysates were collected for protein analysis. Quantification of western blots shows that forced overexpression of miR-34a, miR-34b, or miR-34c suppressed PDL1 protein expression compared with a scrambled control. K) Luciferase activity in cells cotransfected with miR-34a, -b, or -c or a scrambled control and a luciferase reporter construct encoding the luciferase gene fused either to the wild-type PDL1 3’ UTR (PDL1 wt) or a mutated PDL1 3’ UTR (PDL1 mut). All three of the miR-34s reduced luciferase activity (_P < ._001, P =.006, and P =.006). An unpaired t test was used to calculate the two-sided P values. *P < .05, **P < .01, ***P < .001. Error bars on the bar charts represent standard deviation. Similar results were observed in three replicates. mut = mutated; NSCLC = non–small cell lung cancer; scr = scrambled; wt = wild-type.
Figure 4.
PDL1 expression after in vivo delivery of miR-34a. Analysis of miR-34a and PDL1 expression levels in subcutaneous 344SQ tumors treated with MRX34 (n = 2) by quantitative polymerase chain reaction (A), by western blotting (C), by flow cytometry (B) (P = .04, n = 2; an unpaired t test was used to calculate the two-sided P value) (D), or by immunohistochemical staining (E) in a syngeneic mouse model. Scale bar = 200 μm. F) MRX34-induced downregulation of PDL1 in an H1299 xenograft model described elsewhere (37). Magnification ×100. Scale bar = 100 μm. *P < .05. Error bars on the bar charts represent standard deviation. ctrl = control; IHC = immunohistochemistry; MRX34 = miR-34a–loaded liposomes.
Figure 5.
Impact of therapeutic miR-34a delivery combined with radiotherapy on immune cell populations in the tumor microenvironment. Subcutaneous tumors were created by inoculating 1 x 106 344SQ cells derived from a spontaneous subcutaneous lung metastasis from a p53R172HΔg/+K-rasLA1/+ mouse (25) into the right leg of each syngeneic 129Sv/Ev mouse. One week after tumor implantation, mice were randomly assigned to one of four groups: control, MRX34 only, radiation (XRT), or MRX34 plus XRT. The MRX formulation was given as subcutaneous injections at a dose of 1mg/kg (total of 8 injections), and local irradiation was given in 6-Gy fractions to a total dose of 18 Gy over three days, starting when the tumors were 8mm in diameter. For the combination-therapy condition, MRX34 was given one hour before XRT. A) One week after treatment completion, flow cytometry revealed that MRX34 + XRT increased the number of CD8+ cells compared with control or either treatment given alone (P = .004, n = 4). B and C) MRX34 reduced the numbers of PD1+ T-cells (B) and macrophages (C) (P = .001 and P = .008, n = 4). MRX34+XRT combination treatment was more effective in reducing the numbers of PD1+ T-cells (P = .02, n = 4) and macrophages than was XRT alone. (d) XRT seemed to increase the numbers of T-regulatory cells (Tregs) over the control condition, but none of the treatments showed statistically significant effects on Tregs. E) Interferon-gamma levels were increased by MRX34 only and by MRX34 + XRT vs control or XRT alone; MRX34 + XRT increased levels of tumor necrosis factor–alpha. An unpaired t test was used to calculate the two-sided P values. F) MRX34 + XRT delayed tumor growth compared with the control condition in a 344SQ mouse model (n = 6). *P < .05. Error bars on the bar charts represent standard deviation. IFNγ = interferon-gamma; MRX34 = miR-34a–loaded liposomes; TNFα = tumor necrosis factor–alpha; XRT = radiotherapy.
Similar articles
- Acacetin inhibited non-small-cell lung cancer (NSCLC) cell growth via upregulating miR-34a in vitro and in vivo.
Li J, Zhong X, Zhao Y, Shen J, Xiao Z, Pilapong C. Li J, et al. Sci Rep. 2024 Jan 29;14(1):2348. doi: 10.1038/s41598-024-52896-6. Sci Rep. 2024. PMID: 38287075 Free PMC article. - Luteolin Inhibits Tumorigenesis and Induces Apoptosis of Non-Small Cell Lung Cancer Cells via Regulation of MicroRNA-34a-5p.
Jiang ZQ, Li MH, Qin YM, Jiang HY, Zhang X, Wu MH. Jiang ZQ, et al. Int J Mol Sci. 2018 Feb 2;19(2):447. doi: 10.3390/ijms19020447. Int J Mol Sci. 2018. PMID: 29393891 Free PMC article. - CircKRT1 drives tumor progression and immune evasion in oral squamous cell carcinoma by sponging miR-495-3p to regulate PDL1 expression.
Yang Z, Chen W, Wang Y, Qin M, Ji Y. Yang Z, et al. Cell Biol Int. 2021 Jul;45(7):1423-1435. doi: 10.1002/cbin.11581. Epub 2021 Apr 14. Cell Biol Int. 2021. PMID: 33675276 - Emerging role of tumor suppressing microRNAs as therapeutics in managing non-small cell lung cancer.
Singh S, Saxena S, Sharma H, Paudel KR, Chakraborty A, MacLoughlin R, Oliver BG, Gupta G, Negi P, Singh SK, Dua K. Singh S, et al. Pathol Res Pract. 2024 Apr;256:155222. doi: 10.1016/j.prp.2024.155222. Epub 2024 Feb 23. Pathol Res Pract. 2024. PMID: 38452582 Review. - The guardian's little helper: microRNAs in the p53 tumor suppressor network.
He X, He L, Hannon GJ. He X, et al. Cancer Res. 2007 Dec 1;67(23):11099-101. doi: 10.1158/0008-5472.CAN-07-2672. Cancer Res. 2007. PMID: 18056431 Review.
Cited by
- Prognostic and Predictive Biomarkers in Non-Small Cell Lung Cancer Patients on Immunotherapy-The Role of Liquid Biopsy in Unraveling the Puzzle.
Augustus E, Zwaenepoel K, Siozopoulou V, Raskin J, Jordaens S, Baggerman G, Sorber L, Roeyen G, Peeters M, Pauwels P. Augustus E, et al. Cancers (Basel). 2021 Apr 2;13(7):1675. doi: 10.3390/cancers13071675. Cancers (Basel). 2021. PMID: 33918147 Free PMC article. Review. - Predicting therapeutic responses in head and neck squamous cell carcinoma from TP53 mutation detected by cell-free DNA.
Wei M, Zhi J, Li L, Wang W. Wei M, et al. Transl Cancer Res. 2023 Dec 31;12(12):3604-3617. doi: 10.21037/tcr-23-878. Epub 2023 Dec 11. Transl Cancer Res. 2023. PMID: 38197078 Free PMC article. - Influence of TP53 Comutation on the Tumor Immune Microenvironment and Clinical Outcomes With Immune Checkpoint Inhibitors in _STK11_-Mutant Non-Small-Cell Lung Cancer.
Naqash AR, Floudas CS, Aber E, Maoz A, Nassar AH, Adib E, Choucair K, Xiu J, Baca Y, Ricciuti B, Alessi JV, Awad MM, Kim C, Judd J, Raez LE, Lopes G, Nieva JJ, Borghaei H, Takebe N, Ma PC, Halmos B, Kwiatkowski DJ, Liu SV, Mamdani H. Naqash AR, et al. JCO Precis Oncol. 2024 Feb;8:e2300371. doi: 10.1200/PO.23.00371. JCO Precis Oncol. 2024. PMID: 38330261 Free PMC article. - Global burden, risk factors, clinicopathological characteristics, molecular biomarkers and outcomes of microsatellite instability-high gastric cancer.
Zhang Z, Huang J, Li Y, Yan H, Xie J, Wang J, Zhao B. Zhang Z, et al. Aging (Albany NY). 2024 Jan 12;16(1):948-963. doi: 10.18632/aging.205431. Epub 2024 Jan 12. Aging (Albany NY). 2024. PMID: 38224334 Free PMC article. Review. - Co-expression of PD-L1 and HIF-1α predicts poor prognosis in Patients with Non-small Cell Lung Cancer after surgery.
Zheng H, Ning Y, Zhan Y, Liu S, Yang Y, Wen Q, Fan S. Zheng H, et al. J Cancer. 2021 Feb 2;12(7):2065-2072. doi: 10.7150/jca.53119. eCollection 2021. J Cancer. 2021. PMID: 33754005 Free PMC article.
References
- Pfeifer GP, Holmquist GP. Mutagenesis in the P53 gene. Biochim Biophys Acta. 1997;1333 (1):M1–M8. - PubMed
- Elias J, Dimitrio L, Clairambault J, et al. The p53 protein and its molecular network: modelling a missing link between DNA damage and cell fate. Biochim Biophys Acta. 2014;1844(1 Pt B):232–247. - PubMed
- Rufini A, Tucci P, Celardo I, et al. Senescence and aging: the critical roles of p53. Oncogene. 2013;32 (43):5129–5143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- 1 R01 CA182905-01/CA/NCI NIH HHS/United States
- 1UH2TR00943-01/TR/NCATS NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous