Structural basis for the binding of tryptophan-based motifs by δ-COP - PubMed (original) (raw)
Structural basis for the binding of tryptophan-based motifs by δ-COP
Richard J Suckling et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Coatomer consists of two subcomplexes: the membrane-targeting, ADP ribosylation factor 1 (Arf1):GTP-binding βγδζ-COP F-subcomplex, which is related to the adaptor protein (AP) clathrin adaptors, and the cargo-binding αβ'ε-COP B-subcomplex. We present the structure of the C-terminal μ-homology domain of the yeast δ-COP subunit in complex with the WxW motif from its binding partner, the endoplasmic reticulum-localized Dsl1 tether. The motif binds at a site distinct from that used by the homologous AP μ subunits to bind YxxΦ cargo motifs with its two tryptophan residues sitting in compatible pockets. We also show that the Saccharomyces cerevisiae Arf GTPase-activating protein (GAP) homolog Gcs1p uses a related WxxF motif at its extreme C terminus to bind to δ-COP at the same site in the same way. Mutations designed on the basis of the structure in conjunction with isothermal titration calorimetry confirm the mode of binding and show that mammalian δ-COP binds related tryptophan-based motifs such as that from ArfGAP1 in a similar manner. We conclude that δ-COP subunits bind Wxn(1-6)[WF] motifs within unstructured regions of proteins that influence the lifecycle of COPI-coated vesicles; this conclusion is supported by the observation that, in the context of a sensitizing domain deletion in Dsl1p, mutating the tryptophan-based motif-binding site in yeast causes defects in both growth and carboxypeptidase Y trafficking/processing.
Keywords: COPI; coatomer; membrane trafficking; vesicle coat; δ-COP μ homology domain-binding motifs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
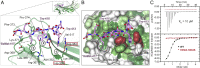
Fig. 1.
Structure of yeast δ-COP MHD with the yeast Dsl1p WxWxϕ motif. (A) Schematic representation of interactions between δ-COP and Dsl1p. (B) ITC experiments showed that yeast δ-COP MHD (residues 282–546) binds to di-tryptophan peptides corresponding to the WxW (DDW413NW415EVED) and WxxxW (ENAW455DEAW459AIDEC) motifs in yeast Dsl1p with K_Ds of 11 ± 2 and 16 ± 1 μM (SD, n = 3) respectively, at a 1:1 stoichiometric ratio. (C) Crystal structure of yeast δ-COP MHD with a SeMet-substituted Dsl1p WxW peptide [DDWNWE(SeMet)ED] showing that di-tryptophan motifs bind to δ-COP at a position similar to that at which the YKFFE sequence of APP binds to the μ4 MHD (PDB ID code 3L81) (17) but at a completely different site from the binding of Yxxϕ signals to AP MHDs (PDB ID code 1BXX) (16, 18, 19). The final refined 2_mF O -DF C density contoured at 1.2σ and mF O -DF C difference density at ±3.0σ for the peptide are shown in the Inset. The total buried area of the interface between δ-COP and the WxWx(SeMet) peptide is 1,354 Å2, as calculated by the PISA server (
). (D) The Consurf server (
consurf.tau.ac.il/
) was used to create a surface representation of the evolutionary conservation of residues in δ-COP MHD based on an alignment from yeast to humans using ClustalO (see also Fig. S3). The tryptophan-based motif-binding site is the outstanding feature of this conservation surface representation. Surface representations of the electrostatic potential of δ-COP MHD and μ2 MHD, highlight that δ-COP MHD is largely negative [contoured from −0.5 V (red) to +0.5 V (blue)]. The di-tryptophan motif-binding site on δ-COP MHD is relatively positive compared with the remainder of the domain. The conservation surface representation shows that the negatively charged surface on the backside of subdomain B is also highly conserved.
Fig. S1.
δ-COP MHD binds to W(x)nW motifs where n = 4, 5, or 6. (A) ITC experiments showed that yeast δ-COP MHD (residues 282–546) (W404A) binds to W(x)nW peptides (TDDGWDNQNW, TDDGWDNSQNW, and TDDGWDNSSQNW) where n = 4, 5, or 6 with _K_Ds of 10, 21 ± 2, and 38 ± 2 μM (SD, n = 3), respectively, at a 1:1 stoichiometric ratio. (B) ITC showing that δ-COP MHD (residues 282–546) (W404A) binds to a peptide [DDWNWE(SeMet)ED] corresponding to the WxW motif in the Dsl1p lasso, in which the valine in the native sequence was replaced by a SeMet to help identify the peptide orientation. The peptide bound with an affinity similar to that of the native peptide [K_d = 4 ± 1 μM vs. 11 ± 2 μM (SD); n = 3)] (Fig. 1_B) at a 1:1 stoichiometric ratio. The slightly higher affinity of the SeMet-derived peptide presumably results from additional van der Waals interactions of the methionine side chain with hydrophobic pocket 3, as compared with the native valine side chain (Fig. S2). (C) ITC showed that a combination of H330A and K363S mutations in human δ-COP (homologous to the H350A and R384S mutations in yeast δ-COP; see Fig. S3) abolishes (_K_d >300 μM) the interaction of the human ArfGAP1 WxxxxW peptide (TDDGWDNQNW) (red squares) with δ-COP MHD (residues 272–511) (W381A) (black squares). The lack of peptide binding shown by the H330A/K363S shows that the human ArfGAP1 WxxxxW peptide (TDDGWDNQNW) binds to the human δ-COP MHD at the same site as the Gcs1p WxxF peptide binds to the yeast δ-COP MHD. Data for the H330A K363S W381A mutant are offset by +0.25 μcal/s for clarity.
Fig. S2.
Structural overview of δ-COP MHD binding to tryptophan-based motifs: electron density from δ-COP MHD in complex with the Dsl1p WxWx(SeMet) peptide (A_–_C), with the Dsl1p WxxxW peptide (D and E), with the Dsl1p WxWxV native peptide (F and G), and with the Gcs1p WxxF peptide (H and I). (A_–_C) Part of the experimentally phased, solvent-flattened electron density map from autoSHARP (44) highlighting the clear protein–solvent boundary (A and B) and the clear density for the peptide (B and C) (1.0σ). In C an anomalous difference map (solid) (±3.0σ; yellow = positive; red = negative) (clipped around peptide) shows the large positive peak representing the SeMet in the WxWx(SeMet) peptide. (D and E) 2mFO-FC (0.8σ) and mFO-FC (±2.5σ) density maps representing the WxxxW peptide before (D) and after (E) peptide model building, showing that the peptide occupancy is low, and the density is difficult to interpret. However the peptide most likely binds in the same orientation as the other peptides. (F and G) 2mFO-FC (0.8σ) and mFO-FC (±3.0σ) density maps representing the native Dsl1p WxWxV peptide before (F) and after (G) peptide model building, showing the clear density for the two tryptophans. (H and I) 2mFO-FC (1.0σ) and mFO-FC (±3.0σ) density maps representing the Gcs1p WxxF peptide before (H) and after (I) peptide model building, showing the clear density for the tryptophan and phenylalanine.
Fig. S3.
Conservation of the tryptophan-based motif-binding site in δ-COP MHD. The Consurf server (
consurf.tau.ac.il/
) was used to create a surface representation of evolutionary conservation of residues in the δ-COP MHD (also shown in Fig. 1_D_), based on an alignment from yeast to humans using ClustalO (shown here). The region that forms the tryptophan-based motif-binding site in this alignment is shown here. Residues directly involved in the binding site are highlighted above the alignment in red. The tryptophan-based motif-binding site is the outstanding feature of the conservation surface representation, indicating its importance. The Dsl1p WxW peptide is shown. Note the conservation of the length of the loop between strands 5 and 6.
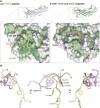
Fig. 2.
Structural details of Dsl1p WxW peptide bound to δ-COP MHD. (A) The Dsl1p WxWx(SeMet) peptide [DDWNWE(SeMet)ED] binds to the δ-COP MHD in an extended conformation with the three hydrophobic residues binding into three complementary pockets (labeled). (B) Surface representation of the di-tryptophan motif-binding site in the δ-COP MHD colored from high (dark green) to low (white) hydrophobicity. The three hydrophobic pockets into which the hydrophobic residues in the peptide bind can be seen clearly. (C) ITC showing that a combination of H350A and R384S mutations (highlighted in dark red in A and B) abolish (_K_d >300 μM) (red triangles) the interaction of the Dsl1p WxW peptide (DDWNWEVED) with δ-COP MHD (black squares) (_K_d = 11 ± 2 μM) (SD, n = 3). Data for the mutant are offset for clarity.
Fig. S4.
The δ-COP MHD binds to tryptophan-based motifs at a site similar to that at which μ4 binds to the YKFFE motif. (A) Surface representations of the YKFFE-binding site in the μ4 MHD (PDB ID code 3L81) (17) and the tryptophan-based motif-binding site in δ-COP MHD colored from high (dark green) to low (white) hydrophobicity with the relevant peptides shown. This representation shows that pocket 1 (the pocket between His-350 and Arg-384) in δ-COP is equivalent to the pocket in μ4 in which Phe-690 (YKFFE) in the APP motif is buried, as is evident from the superposition of the μ4 and δ-COP MHDs. (B) Three different angles of the superposed peptides are shown.
Fig. S5.
Binding of tryptophan-based motifs to the δ-COP MHD through a lock-and-key mechanism. Superposition of the δ-COP MHD bound to peptide (dark green) [of the δ-COP: Dsl1p WxWx(SeMet) peptide structure] with a molecule not bound to peptide (light green) shows that the binding of peptide has no discernable effect on the structure. The only clear difference is the different conformation of the Leu-380 side chain, creating a larger hydrophobic pocket into which the second tryptophan binds (pocket 2).
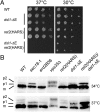
Fig. 3.
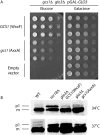
ret2(HARS)/dsl1-ΔE mutant cells show defects in growth and in CPY trafficking. (A) Isogenic strains bearing mutations at either the DSL1 or RET2 (δ-COP) gene loci, labeled as dsl1-ΔE and ret2(HARS), were serially diluted, spotted, and grown at the indicated temperatures for 48 h. The double mutant harboring both the ret2(HARS) and dsl1-ΔE allele was lethal at 37 °C (no colonies formed after 1 wk) but was viable at lower temperatures, although with a slight growth defect. All combinations are shown in biological duplicate. (B) Cells of the indicated strains were grown to midlog at 30 °C in yeast extract peptone dextrose (YEPD) medium and then were shifted for 2 h to either 34 °C or 37 °C. Glass bead extracts of collected cells were separated by 7.5% SDS-PAGE and probed with α-CPY antibodies (40). Positions of p1 (ER), p2 (Golgi), and mature (m, vacuolar) forms of CPY are indicated. Note that p1 CPY accumulates in ret2(HARS) dsl1-ΔE double-mutant cells at 34 °C and more strongly at 37 °C but does not accumulate in single-mutant cells.
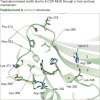
Fig. 4.
Structural details of Gcs1p WxxF peptide bound to δ-COP MHD. (A, Left) Alignment of the C-terminal region of Gcs1p from different yeasts showing that the WxxF motif at the extreme C terminus is conserved in all except S. pombe. (Right) δ-COP in S. pombe does not contain an MHD; the lack of selective pressure means the WxxF in the Gcs1p homolog in S. pombe has been lost. (B) Gcs1p WxxF peptide (DEDKWDDF) binds to δ-COP MHD at the same site where the Dsl1p WxW peptide binds with Trp-349 in pocket 1 and Phe-352 in pocket 2. (C) ITC showing that a combination of H350A and R384S mutations (highlighted in dark red in B) also abolish (_K_d >300 μM) (red triangles) the interaction of the Gcs1p WxxF peptide (DEDKWDDF) with the δ-COP MHD (black squares) (_K_d = 11 ± 1 μM at a 1:1 stoichiometric ratio; n = 3). Data for mutant are offset for clarity. (D) Superposition of the two δ-COP MHD tryptophan-based peptide cocrystal structures showing that the N-terminal tryptophan of both peptides binds into pocket 1, and the C-terminal tryptophan/phenylalanine into pocket 2. Superposition is within the tryptophan motif-binding site (residues 348–355 and 363–384). For clarity only the model of δ-COP is shown from the Gcs1p WxxF peptide cocrystal structure, and only the hydrophobic residue side chains within the peptides are shown.
Fig. S6.
Compromised Gcs1p binding to the δ-COP MHD impairs yeast cell growth. (A) Yeast cells carrying simultaneous chromosomal deletions of the ArfGAP genes GCS1 and GLO3 are not viable unless the expression of one of these ArfGAPs is driven from a plasmid. Galactose-dependent expression of Glo3p from the GAL10 promoter can keep double-mutant cells growing in medium containing galactose but not in galactose-free (i.e., glucose-containing) medium. Individual plasmid transformants were serially diluted, spotted onto glucose-containing plates, and incubated at 30 °C for 48 h. Note that the mutant Gcs1-AA protein (with the W349A, F352A substitutions) cannot sustain optimal yeast colony growth in galactose-free medium, in comparison with and in contrast to wild-type Gcs1p. (B) Cells of the indicated strains (the RET2 gene encodes δ-COP) were grown to midlog phase in YEPD medium at 30 °C and then were shifted to 34 °C or 37 °C for 2 h. Glass bead extracts of collected cells were separated by 7.5% SDS-PAGE and were probed with anti-CPY antibodies (see ref. 40). Positions of p1 (ER) and mature (m, vacuolar) forms of CPY are indicated. glo3Δ/_gcs1_-WF single- and the _glo3_Δ/_gcs1_-AA double-mutant cells display a strong defect in ER-to-Golgi transport of CPY at 37 °C but not at 34 °C. Note that the mutants do not differ noticeably in their CPY-processing ability. Extensive attempts to demonstrate the physical interaction between coatomer and Gcs1p in vivo have proved technically impossible, because the levels of detergent needed to reduce nonspecific background binding to acceptable levels [>1% (vol/vol) IGEPAL; Sigma-Aldrich] abrogate the interaction in vitro. This lack of binding in the presence of <1% IGEPAL likely indicates that the interaction is mediated mainly by hydrophobic side-chain interactions that are readily outcompeted by detergent molecules.
Fig. S7.
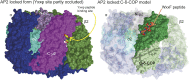
Docking of the δ-COP MHD structure into the AP2 locked/closed form. Surface representation of the AP2 locked form (PDB ID code 2VGL) (21) and docking of the δ-COP MHD (C-δ-COP)-Gcs1p WxxF peptide cocrystal structure into the AP2 locked form based on the superposition of C-δ-COP with C-μ2. The Gcs1p WxxF peptide is shown as spheres. Note the significant occlusion of the Yxxϕ binding site in μ2 in the AP2 structure and the fully accessible WxxF binding site in δ-COP.
Similar articles
- Rules for the recognition of dilysine retrieval motifs by coatomer.
Ma W, Goldberg J. Ma W, et al. EMBO J. 2013 Apr 3;32(7):926-37. doi: 10.1038/emboj.2013.41. Epub 2013 Mar 12. EMBO J. 2013. PMID: 23481256 Free PMC article. - Mutational analysis of betaCOP (Sec26p) identifies an appendage domain critical for function.
DeRegis CJ, Rahl PB, Hoffman GR, Cerione RA, Collins RN. DeRegis CJ, et al. BMC Cell Biol. 2008 Jan 22;9:3. doi: 10.1186/1471-2121-9-3. BMC Cell Biol. 2008. PMID: 18211691 Free PMC article. - The structure of COPI vesicles and regulation of vesicle turnover.
Taylor RJ, Tagiltsev G, Briggs JAG. Taylor RJ, et al. FEBS Lett. 2023 Mar;597(6):819-835. doi: 10.1002/1873-3468.14560. Epub 2022 Dec 30. FEBS Lett. 2023. PMID: 36513395 Review. - The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants.
Memon AR. Memon AR. Biochim Biophys Acta. 2004 Jul 1;1664(1):9-30. doi: 10.1016/j.bbamem.2004.04.005. Biochim Biophys Acta. 2004. PMID: 15238254 Review.
Cited by
- ER arrival sites for COPI vesicles localize to hotspots of membrane trafficking.
Schröter S, Beckmann S, Schmitt HD. Schröter S, et al. EMBO J. 2016 Sep 1;35(17):1935-55. doi: 10.15252/embj.201592873. Epub 2016 Jul 20. EMBO J. 2016. PMID: 27440402 Free PMC article. - COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal.
Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR. Xu P, et al. Elife. 2017 Oct 23;6:e28342. doi: 10.7554/eLife.28342. Elife. 2017. PMID: 29058666 Free PMC article. - ARF GTPases and their GEFs and GAPs: concepts and challenges.
Sztul E, Chen PW, Casanova JE, Cherfils J, Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schürmann A, Wilhelmi I, Yohe ME, Kahn RA. Sztul E, et al. Mol Biol Cell. 2019 May 15;30(11):1249-1271. doi: 10.1091/mbc.E18-12-0820. Mol Biol Cell. 2019. PMID: 31084567 Free PMC article. Review. - COPB2 gene silencing inhibits colorectal cancer cell proliferation and induces apoptosis via the JNK/c-Jun signaling pathway.
Wang Y, Xie G, Li M, Du J, Wang M. Wang Y, et al. PLoS One. 2020 Nov 19;15(11):e0240106. doi: 10.1371/journal.pone.0240106. eCollection 2020. PLoS One. 2020. PMID: 33211699 Free PMC article. - Structural characterization of coatomer in its cytosolic state.
Wang S, Zhai Y, Pang X, Niu T, Ding YH, Dong MQ, Hsu VW, Sun Z, Sun F. Wang S, et al. Protein Cell. 2016 Aug;7(8):586-600. doi: 10.1007/s13238-016-0296-z. Epub 2016 Jul 29. Protein Cell. 2016. PMID: 27472951 Free PMC article.
References
- Szul T, Sztul E. COPII and COPI traffic at the ER-Golgi interface. Physiology (Bethesda) 2011;26(5):348–364. - PubMed
- Presley JF, et al. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417(6885):187–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105178845/MRC_/Medical Research Council/United Kingdom
- U105178845/MRC_/Medical Research Council/United Kingdom
- CAPMC/ CIHR/Canada
- 090909/WT_/Wellcome Trust/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- T32 GM007388/GM/NIGMS NIH HHS/United States
- R01 GM071574/GM/NIGMS NIH HHS/United States
- GM071574/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous