Serial interactome capture of the human cell nucleus - PubMed (original) (raw)
Serial interactome capture of the human cell nucleus
Thomas Conrad et al. Nat Commun. 2016.
Abstract
Novel RNA-guided cellular functions are paralleled by an increasing number of RNA-binding proteins (RBPs). Here we present 'serial RNA interactome capture' (serIC), a multiple purification procedure of ultraviolet-crosslinked poly(A)-RNA-protein complexes that enables global RBP detection with high specificity. We apply serIC to the nuclei of proliferating K562 cells to obtain the first human nuclear RNA interactome. The domain composition of the 382 identified nuclear RBPs markedly differs from previous IC experiments, including few factors without known RNA-binding domains that are in good agreement with computationally predicted RNA binding. serIC extends the number of DNA-RNA-binding proteins (DRBPs), and reveals a network of RBPs involved in p53 signalling and double-strand break repair. serIC is an effective tool to couple global RBP capture with additional selection or labelling steps for specific detection of highly purified RBPs.
Figures
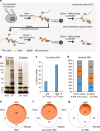
Figure 1. serIC recovers highly purified nuclear RBPs.
(a) Comparison of the IC and serIC methods. Irradiation of living cells with ultraviolet (UV) light at 254 nm induces covalent crosslinks between proteins and nucleic acids. After cell lysis, RBP–poly(A)-RNA complexes are captured by hybridization to oligo(dT) magnetic beads and washed under denaturing conditions. In IC, purified material is eluted and proteins are identified by LC–MS/MS (top). In serIC, eluted material is enzymatically treated to remove contaminating DNA. Free RBP–poly(A)-RNA complexes are diluted in the presence of high salt and detergent to disrupt residual non-crosslinked interactions, recaptured by hybridization to oligo(dT) beads and analysed by LC–MS/MS. (b) SDS–PAGE of proteins crosslinked to poly(A)-RNA. Nuclei from irradiated K562 cells were isolated and subjected to the serIC procedure. Protein–RNA complexes obtained after the first or second round of purification (first elution and second elution) were digested with RNAseA/T1, separated on a polyacrylamide-gradient gel, and visualized by silver staining. No UV-crosslinking was performed in controls (No Cl). Asterisks indicate RNase A/T1. (c) Elimination of DNAse contamination. qPCR measurement of eluate from the first and second round of purification was performed without reverse transcription to detect residual DNA. Levels of 45S rDNA and LINE 1.3 retrotransposon DNA in the first and second eluate are shown relative to GAPDH mRNA in the second eluate. Error bars show s.d. from two independent experiments. (d) Domain composition of the nuclear RNA interactome. Proteins identified by LC–MS/MS in serIC preparations from K562 nuclei, and in previous IC from HeLa and HEK293 (ref. 14) were grouped by the presence or absence of classical or non-classical RBDs as indicated. (e–g) serIC recovery of proteins with the GO association ‘nucleus' that were previously identified by IC in HeLa or HEK293 (ref. 14). Proteins containing classical (e), non-classical (f), or no RBDs (g) were compared separately.
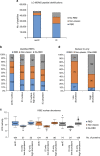
Figure 2. Altered domain composition of the serIC RNA interactome.
(a) The combined number of individual LC–MS/MS peptide identifications in all serIC replicates and in all IC replicates from HeLa. Indicated are peptides from proteins with classic, non-classic or no RBD. Only peptides from proteins that are enriched over the control are shown for each experiment. (b) Domain composition of the nuclear RNA interactome. Proteins identified by IC in HeLa and HEK293 (ref. 14) were filtered for expression in K562 nuclei or cytoplasm. All nuclear, only highly abundant nuclear, and cytoplasmic IC-derived candidates are included in the comparison, respectively. (c) K562 nuclear abundance of the RBPs detected by serIC or in IC only, as measured by LC–MS/MS. IC-only candidates are subdivided into proteins with similar abundance as serIC-derived RBPs from the same class, and proteins with reduced abundance. (d) Distribution of lowly or highly abundant candidate RBPs. Highly abundant IC-only candidates mostly lack RNA-binding domains.
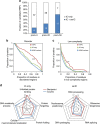
Figure 3. Good agreement between serIC and computational RBP prediction.
(a) All nuclear no-RBD candidate RBPs detected by IC in HEK293, grouped by the precision of computational RNA-binding prediction. Highlighted in blue are the proportion and number of proteins that are also identified as RBP by serIC. (b) Proportion of residues in disordered regions in all K562 nuclear proteins (blue), serIC-derived no-RBD proteins (red) and abundant IC-only nuclear no-RBD candidates (green). (c) Proportion of residues in low complexity regions with colour code as in b. Asterisks in b and c indicate P values <10−3 and<10−2, respectively. (d) GO-enrichment analysis of serIC-derived and IC-only nuclear no-RBD proteins. Shown are the most significantly enriched non-redundant ‘Biological Process' and ‘Molecular Function' GO terms compared with the human genome.
Figure 4. Novel DRBPs connect transcription regulation to the DDR.
All annotated DNA-binding proteins identified in the nuclear RNA interactome. Proteins without classic RBDs were interrogated for previous direct evidence for RNA binding.
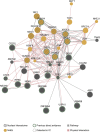
Figure 5. Network of DDR proteins in the nuclear RNA interactome.
Nuclear RNA interactome proteins with GO annotations related to the DDR were used as query to construct a functional interaction network. First, neighbours were identified using the association data from protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. Proteins were grouped based on physical and pathway interactions. Indicated are proteins involved in the NHEJ pathway and proteins with previous evidence for RNA binding.
Similar articles
- The RNA-binding protein repertoire of embryonic stem cells.
Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. Kwon SC, et al. Nat Struct Mol Biol. 2013 Sep;20(9):1122-30. doi: 10.1038/nsmb.2638. Epub 2013 Aug 4. Nat Struct Mol Biol. 2013. PMID: 23912277 - mRNA interactome capture in mammalian cells.
Kastelic N, Landthaler M. Kastelic N, et al. Methods. 2017 Aug 15;126:38-43. doi: 10.1016/j.ymeth.2017.07.006. Epub 2017 Jul 11. Methods. 2017. PMID: 28710009 - Tandem RNA isolation reveals functional rearrangement of RNA-binding proteins on _CDKN1B/p27_Kip1 3'UTRs in cisplatin treated cells.
Iadevaia V, Wouters MD, Kanitz A, Matia-González AM, Laing EE, Gerber AP. Iadevaia V, et al. RNA Biol. 2020 Jan;17(1):33-46. doi: 10.1080/15476286.2019.1662268. Epub 2019 Sep 16. RNA Biol. 2020. PMID: 31522610 Free PMC article. - RNA-Binding Proteins Revisited - The Emerging Arabidopsis mRNA Interactome.
Köster T, Marondedze C, Meyer K, Staiger D. Köster T, et al. Trends Plant Sci. 2017 Jun;22(6):512-526. doi: 10.1016/j.tplants.2017.03.009. Epub 2017 Apr 12. Trends Plant Sci. 2017. PMID: 28412036 Review. - Cooperation and competition by RNA-binding proteins in cancer.
Nag S, Goswami B, Das Mandal S, Ray PS. Nag S, et al. Semin Cancer Biol. 2022 Nov;86(Pt 3):286-297. doi: 10.1016/j.semcancer.2022.02.023. Epub 2022 Mar 3. Semin Cancer Biol. 2022. PMID: 35248729 Review.
Cited by
- Development and Validation of a RNA Binding Protein-Associated Prognostic Model for Hepatocellular Carcinoma.
Wang M, Jiang F, Wei K, Wang J, Zhou G, Wu C, Yin G. Wang M, et al. Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211004936. doi: 10.1177/15330338211004936. Technol Cancer Res Treat. 2021. PMID: 33910445 Free PMC article. - Capture of the newly transcribed RNA interactome using click chemistry.
Guo X, Tariq M, Lai Y, Kanwal S, Lv Y, Wang X, Li N, Jiang M, Meng J, Hu J, Yuan J, Luo Z, Ward C, Volpe G, Wang D, Yin M, Qin B, Zhang B, Bao X, Esteban MA. Guo X, et al. Nat Protoc. 2021 Nov;16(11):5193-5219. doi: 10.1038/s41596-021-00609-y. Epub 2021 Oct 25. Nat Protoc. 2021. PMID: 34697467 - Identification, quantification and bioinformatic analysis of RNA-dependent proteins by RNase treatment and density gradient ultracentrifugation using R-DeeP.
Caudron-Herger M, Wassmer E, Nasa I, Schultz AS, Seiler J, Kettenbach AN, Diederichs S. Caudron-Herger M, et al. Nat Protoc. 2020 Apr;15(4):1338-1370. doi: 10.1038/s41596-019-0261-4. Epub 2020 Feb 24. Nat Protoc. 2020. PMID: 32094787 Free PMC article. - Chemoproteomic capture of RNA binding activity in living cells.
Heindel AJ, Brulet JW, Wang X, Founds MW, Libby AH, Bai DL, Lemke MC, Leace DM, Harris TE, Hafner M, Hsu KL. Heindel AJ, et al. Nat Commun. 2023 Oct 7;14(1):6282. doi: 10.1038/s41467-023-41844-z. Nat Commun. 2023. PMID: 37805600 Free PMC article. - Post-transcriptional regulatory patterns revealed by protein-RNA interactions.
Zanzoni A, Spinelli L, Ribeiro DM, Tartaglia GG, Brun C. Zanzoni A, et al. Sci Rep. 2019 Mar 13;9(1):4302. doi: 10.1038/s41598-019-40939-2. Sci Rep. 2019. PMID: 30867517 Free PMC article.
References
- Fatica A. & Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2014) . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous