A microscopic landscape of the invasive breast cancer genome - PubMed (original) (raw)
A microscopic landscape of the invasive breast cancer genome
Zheng Ping et al. Sci Rep. 2016.
Abstract
Histologic grade is one of the most important microscopic features used to predict the prognosis of invasive breast cancer and may serve as a marker for studying cancer driving genomic abnormalities in vivo. We analyzed whole genome sequencing data from 680 cases of TCGA invasive ductal carcinomas of the breast and correlated them to corresponding pathology information. Ten genetic abnormalities were found to be statistically associated with histologic grade, including three most prevalent cancer driver events, TP53 and PIK3CA mutations and MYC amplification. A distinct genetic interaction among these genomic abnormalities was revealed as measured by the histologic grading score. While TP53 mutation and MYC amplification were synergistic in promoting tumor progression, PIK3CA mutation was found to have alleviated the oncogenic effect of either the TP53 mutation or MYC amplification, and was associated with a significant reduction in mitotic activity in TP53 mutated and/or MYC amplified breast cancer. Furthermore, we discovered that different types of genetic abnormalities (mutation versus amplification) within the same cancer driver gene (PIK3CA or GATA3) were associated with opposite histologic changes in invasive breast cancer. In conclusion, our study suggests that histologic grade may serve as a biomarker to define cancer driving genetic events in vivo.
Figures
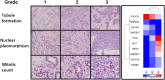
Figure 1. Invasive ductal carcinoma of various Nottingham histological grades associated with driver genomic abnormalities.
Left. Representative microscopic images of invasive ductal carcinoma with various Nottingham histological grades. Red arrow indicates mitotic figure. Right. Hierarchical clustering of 10 candidate breast cancer driver genes correlated to the IDC histologic grades. This was performed using Pearson dissimilarity matrix for both genes and samples. The color indicates sample percentage (%) in G1, G2, and G3 as indicated by the scale.
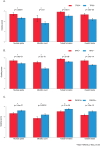
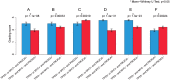
Figure 2. IDC histologic grade in correlation with different states of genetic abnormalities.
(A) Tumors with a TP53 gene mutation, 254 cases; tumors without a TP53 gene mutation, 426 cases. (B) Tumors with MYC gene amplification, 177 cases; tumors without MYC gene amplification, 503 cases. (C) Tumors with PIK3CA gene mutation, 208 cases; tumors without PIK3CA gene mutation, 472 cases. *Indicates Mann-Whitney U test, p < 0.05.
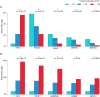
Figure 3
Significant mutated (A) or amplified (B) genes in correlation with histologic grade. *Indicates Fisher exact test, p < 0.05.
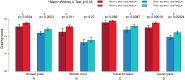
Figure 4. IDC histologic grade in correlation with different states of TP53 mutation and/or MYC amplification.
IDC positive for both TP53 mutation and MYC amplification, 97 cases; IDC positive for TP53 mutation but negative for MYC amplification, 157 cases; IDC positive for MYC amplification but negative for TP53 mutation, 80 cases; IDC negative for both TP53 mutation and MYC amplification, 336 cases. *Indicates Mann-Whitney U test, p < 0.05.
Figure 5. IDC histologic grade in correlation with different states of TP53 mutation, MYC amplification and/or PIK3CA mutation.
IDC positive for TP53 mutation or MYC amplification and also PIK3CA mutation, 81 cases; IDC with TP53 mutation or MYC amplification but negative for PIK3CA mutation, 253 cases; IDC negative for TP53 mutation or MYC amplification but positive for PIK3CA mutation, 127 cases; IDC negative for TP53 mutation, MYC amplification, or PIK3CA mutation, 219 cases. *Indicates Mann-Whitney U test, p < 0.05.
Figure 6. IDC histologic grading features in correlation with various states of TP53 mutation, MYC amplification and/or PIK3CA mutation.
*Indicates Mann-Whitney U test, p < 0.05.
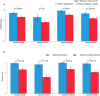
Figure 7
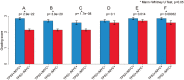
Histologic grade of IDC with amplification or mutation of PIK3CA (A) or GATA3 (B). (A) Tumors with PIK3CA gene mutation, 185 cases; tumors with PIK3CA gene amplification, 46 cases. *Indicates Mann-Whitney U test, p < 0.05. (B) Tumors with GATA3 gene mutation, 67 cases; tumors with GATA3 gene amplification, 37 cases. *Indicates Mann-Whitney U test, p < 0.05.
Similar articles
- Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes.
Kim JY, Lee E, Park K, Park WY, Jung HH, Ahn JS, Im YH, Park YH. Kim JY, et al. Oncotarget. 2017 Apr 25;8(17):27997-28007. doi: 10.18632/oncotarget.15881. Oncotarget. 2017. PMID: 28427202 Free PMC article. - Polarity gene alterations in pure invasive micropapillary carcinomas of the breast.
Gruel N, Benhamo V, Bhalshankar J, Popova T, Fréneaux P, Arnould L, Mariani O, Stern MH, Raynal V, Sastre-Garau X, Rouzier R, Delattre O, Vincent-Salomon A. Gruel N, et al. Breast Cancer Res. 2014 May 8;16(3):R46. doi: 10.1186/bcr3653. Breast Cancer Res. 2014. PMID: 24887297 Free PMC article. - Somatic alterations of TP53, ERBB2, PIK3CA and CCND1 are associated with chemosensitivity for breast cancers.
Yang L, Ye F, Bao L, Zhou X, Wang Z, Hu P, Ouyang N, Li X, Shi Y, Chen G, Xia P, Chui M, Li W, Jia Y, Liu Y, Liu J, Ye J, Zhang Z, Bu H. Yang L, et al. Cancer Sci. 2019 Apr;110(4):1389-1400. doi: 10.1111/cas.13976. Epub 2019 Mar 19. Cancer Sci. 2019. PMID: 30776175 Free PMC article. - p53 in breast cancer subtypes and new insights into response to chemotherapy.
Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A, Bourstyn E, de Cremoux P, Janin A, Giacchetti S, Espié M, de Thé H. Bertheau P, et al. Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review. - TP53 and breast cancer.
Børresen-Dale AL. Børresen-Dale AL. Hum Mutat. 2003 Mar;21(3):292-300. doi: 10.1002/humu.10174. Hum Mutat. 2003. PMID: 12619115 Review.
Cited by
- Exploring the therapeutic mechanisms of Coptidis Rhizoma in gastric precancerous lesions: a network pharmacology approach.
Ye X, Yang C, Xu H, He Q, Sheng L, Lin J, Wang X. Ye X, et al. Discov Oncol. 2024 Jun 5;15(1):211. doi: 10.1007/s12672-024-01070-5. Discov Oncol. 2024. PMID: 38837097 Free PMC article. - Comparison of MUSE-DWI and conventional DWI in the application of invasive breast cancer and malignancy grade prediction: A comparative study.
Wang W, Dou B, Wang Q, Li H, Li C, Zhao W, Fang L, Pylypenko D, Chu Y. Wang W, et al. Heliyon. 2024 Jan 18;10(2):e24379. doi: 10.1016/j.heliyon.2024.e24379. eCollection 2024 Jan 30. Heliyon. 2024. PMID: 38304790 Free PMC article. - A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer.
Amgad M, Hodge JM, Elsebaie MAT, Bodelon C, Puvanesarajah S, Gutman DA, Siziopikou KP, Goldstein JA, Gaudet MM, Teras LR, Cooper LAD. Amgad M, et al. Nat Med. 2024 Jan;30(1):85-97. doi: 10.1038/s41591-023-02643-7. Epub 2023 Nov 27. Nat Med. 2024. PMID: 38012314 - Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel.
Gangapuram M, Mazzio EA, Redda KK, Soliman KFA. Gangapuram M, et al. Int J Mol Sci. 2021 Jul 19;22(14):7694. doi: 10.3390/ijms22147694. Int J Mol Sci. 2021. PMID: 34299315 Free PMC article. - Integrated multi-omics profiling of high-grade estrogen receptor-positive, HER2-negative breast cancer.
Wang K, Li L, Franch-Expósito S, Le X, Tang J, Li Q, Wu Q, Bassaganyas L, Camps J, Zhang X, Li H, Foukakis T, Xiang T, Wu J, Ren G. Wang K, et al. Mol Oncol. 2022 Jun;16(12):2413-2431. doi: 10.1002/1878-0261.13043. Epub 2021 Jul 29. Mol Oncol. 2022. PMID: 34146382 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous