Mechanistic Insights into Archaeal and Human Argonaute Substrate Binding and Cleavage Properties - PubMed (original) (raw)
Mechanistic Insights into Archaeal and Human Argonaute Substrate Binding and Cleavage Properties
Sarah Willkomm et al. PLoS One. 2016.
Abstract
Argonaute (Ago) proteins from all three domains of life are key players in processes that specifically regulate cellular nucleic acid levels. Some of these Ago proteins, among them human Argonaute2 (hAgo2) and Ago from the archaeal organism Methanocaldococcus jannaschii (MjAgo), are able to cleave nucleic acid target strands that are recognised via an Ago-associated complementary guide strand. Here we present an in-depth kinetic side-by-side analysis of hAgo2 and MjAgo guide and target substrate binding as well as target strand cleavage, which enabled us to disclose similarities and differences in the mechanistic pathways as a function of the chemical nature of the substrate. Testing all possible guide-target combinations (i.e. RNA/RNA, RNA/DNA, DNA/RNA and DNA/DNA) with both Ago variants we demonstrate that the molecular mechanism of substrate association is highly conserved among archaeal-eukaryotic Argonautes. Furthermore, we show that hAgo2 binds RNA and DNA guide strands in the same fashion. On the other hand, despite striking homology between the two Ago variants, MjAgo cannot orientate guide RNA substrates in a way that allows interaction with the target DNA in a cleavage-compatible orientation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
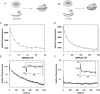
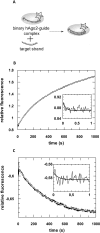
Fig 1. Formation of binary MjAgo-nucleic acid complexes.
Binding of either fluorescently labelled (A) ss guide DNA or (B) ds siDNA to MjAgo was monitored under (C, D) equilibrium or (E, F) pre-steady state conditions. (C) ss guide DNA (D-as2bFAM, 20 nM) or (D) ds siDNA (D-si2bFAM, 20 nM) were titrated with increasing concentrations of MjAgo. Both data sets were mathematically evaluated using a quadratic equation. The best fit of the experimental data to a quadratic equation is shown for representative measurements. The fit yielded K D values of 3.1 (± 0.9) nM and 102 (± 6.6) nM for the binary interaction of MjAgo with ss guide DNA and ds siDNA. Next, binary complex formation was analysed by rapidly mixing (E) 600 nM or (F) 700 nM MjAgo with (E) 20 nM ss guide DNA (D-as2bFAM) or (F) ds siDNA (D-si2bFAM). Representative graphs are shown. The inset shows the data on a shorter time scale. Data were fitted best using a triple exponential equation, yielding the following rate constants: (E) _k_1_obs: 25. 0 (± 6.2) s-1, _k_2: 0.23 (± 0.02) s-1 and _k_3: 0.003 (± 0.0002) s-1 and (F) _k_1_obs: 22.1 (± 1.5) s-1, _k_2: 0.24 (± 0.005) s-1 and _k_3: 0.003 (± 0.0002) s-1.
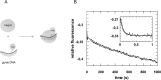
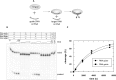
Fig 2. Pre-steady state kinetics of binary hAgo2-guideDNA complex formation.
(A) Schematic representation of the conducted experiment. (B) Binary complex formation was analysed by rapidly mixing 500 nM hAgo2 with 20 nM ss guide DNA (D-as2bFAM). A representative graph is shown. The inset shows the data on a shorter time scale. Data were fitted best using a triple exponential equation yielding the following rate constants: _k_1_obs: 10.6 (± 0.4) s-1, _k_2: 0.11 (± 0.01) s-1 and _k_3: 0.004 (± 0.0003) s-1.
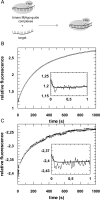
Fig 3. Association of ternary MjAgo-guide-target complexes.
(A) Schematic representation of the experimental setup; MjAgo is pre-assembled with a fluorescently labelled DNA guide and subsequently mixed with an unlabelled guide-matching target (DNA or RNA) strand. Pre-assembled binary complexes consisting of 500 nM MjAgo and 20 nM guide DNA (D-as2bFAM) are rapidly mixed with either (B) 20 nM target DNA (D-s2b) or (C) 40 nM target RNA (s2b). Representative graphs are shown. The insets show the data on a shorter time scale. Data could be fitted best using a triple exponential equation, yielding the following rate constants: (B) _k_1_obs: 21.8 (± 3.0) s-1, _k_2: 0.01 (± 0.0003) s-1 and _k_3: 0.003 (± 0.00007) s-1 or (C) _k_1_obs: 25.1 (± 10.5) s-1, _k_2: 0.04 (± 0.0005) s-1 and _k_3: 0.004 (± 0.00001) s-1.
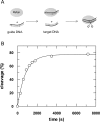
Fig 4. MjAgo-mediated target DNA cleavage under single turnover conditions.
(A) Schematic representation of the experimental setup. MjAgo was premixed with guide DNA. Cleavage was started by adding a radioactively labelled DNA target strand. Cleavage experiments were conducted using 1 μM MjAgo, 100 nM guide strand (D-as2b) and 2.5 nM target strand (32P-D-s2b) at 85°C. Samples were taken at time points 0’, 5’, 10’, 15’, 20’, 25’, 30’, 60’ and 120’, separated on 20% denaturing polyacrylamide gel and visualized by autoradiography. Relative cleavage amplitudes were plotted versus time. Data could be fitted best using a single exponential equation yielding a rate constant k cleavage: 0.0012 (± 0.00007) s-1. The average of three independent measurements yielded a rate constant of 0.0014 (± 0.0003) s-1.
Fig 5. Association of different ternary hAgo2-guide-target complexes.
(A) Schematic representation of the experimental setup; binary complexes composed of hAgo2 and a fluorescently labelled guide strand (RNA or DNA) are preassembled and subsequently mixed with a matching target strand (DNA or RNA). Preassembled binary complexes consisting of 500 nM hAgo2 and 20 nM (B) guide RNA (as2bFAM) or (C) guide DNA (D-as2b) are rapidly mixed with 20 nM (B) target DNA (D-s2b) or (C) target RNA (s2bFAM). Representative graphs are shown. The inset shows the data on a shorter time scale. Data could be fitted best using a triple exponential equation, yielding the following rate constants: (B) _k_1_obs: 9.2 (± 0.9) s-1, _k_2: 0.005 (± 0.00009) s-1 and _k_3: 0.0007 (± 0.00004) s-1 and (C) _k_1_obs: 7.9 (± 0.8) s-1, _k_2: 0.004 (± 0.0004) s-1 and _k_3: 0.001 (± 0.0001) s-1.
Fig 6. hAgo2 cleavage assay with RNA or DNA targets guided by RNA or DNA.
(A) Cleavage experiments were conducted using 2.5 μM hAgo2, 100 nM guide strand and 2.5 nM target strand at 37°C. Samples were taken at time points 0’, 10’, 30’, 60’ and 120’, separated on 20% denaturing polyacrylamide gel and visualised by autoradiography. (B) Bands were evaluated using ImageQuant 5 and relative cleavage amplitudes were plotted versus time. (C) Data could be fitted best with a single exponential equation yielding a rate constant k cleavage: 0.0003 s-1 for RNA cleavage irrespective of the guide strand chemistry. The average of two independent measurements yields a rate constant of 0.0003 (± 0.00007) s-1 in both cases. In case of DNA targets no cleavage could be observed.
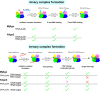
Fig 7. Schematic representation of the nucleic acid binding properties of hAgo2 and MjAgo.
Ago is represented in cartoons based on X-ray structures with N-, PAZ, Mid and PIWI domain coloured individually (N-terminal domain: blue; PAZ: magenta; Mid: yellow; PIWI: green). Relative positions of Ago, guide and target strand are indicated. The corresponding rate constants for each step are given in Table 2. Green checkmarks indicate the individual step is taking place, red crosses show that the indicated step could not be observed. The question mark indicates that this transition could not be definitely assigned.
Similar articles
- Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii.
Zander A, Willkomm S, Ofer S, van Wolferen M, Egert L, Buchmeier S, Stöckl S, Tinnefeld P, Schneider S, Klingl A, Albers SV, Werner F, Grohmann D. Zander A, et al. Nat Microbiol. 2017 Mar 20;2:17034. doi: 10.1038/nmicrobiol.2017.34. Nat Microbiol. 2017. PMID: 28319081 Free PMC article. - Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Willkomm S, Oellig CA, Zander A, Restle T, Keegan R, Grohmann D, Schneider S. Willkomm S, et al. Nat Microbiol. 2017 Mar 20;2:17035. doi: 10.1038/nmicrobiol.2017.35. Nat Microbiol. 2017. PMID: 28319084 - Single-molecule FRET supports the two-state model of Argonaute action.
Zander A, Holzmeister P, Klose D, Tinnefeld P, Grohmann D. Zander A, et al. RNA Biol. 2014;11(1):45-56. doi: 10.4161/rna.27446. Epub 2013 Dec 20. RNA Biol. 2014. PMID: 24442234 Free PMC article. - DNA silencing by prokaryotic Argonaute proteins adds a new layer of defense against invading nucleic acids.
Willkomm S, Makarova KS, Grohmann D. Willkomm S, et al. FEMS Microbiol Rev. 2018 May 1;42(3):376-387. doi: 10.1093/femsre/fuy010. FEMS Microbiol Rev. 2018. PMID: 29579258 Free PMC article. Review. - Conformational Dynamics of Ago-Mediated Silencing Processes.
Willkomm S, Restle T. Willkomm S, et al. Int J Mol Sci. 2015 Jul 1;16(7):14769-85. doi: 10.3390/ijms160714769. Int J Mol Sci. 2015. PMID: 26140373 Free PMC article. Review.
Cited by
- Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA.
Dayeh DM, Cantara WA, Kitzrow JP, Musier-Forsyth K, Nakanishi K. Dayeh DM, et al. Nucleic Acids Res. 2018 Sep 19;46(16):e98. doi: 10.1093/nar/gky496. Nucleic Acids Res. 2018. PMID: 29897478 Free PMC article. - Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence.
Koonin EV. Koonin EV. Biol Direct. 2017 Feb 10;12(1):5. doi: 10.1186/s13062-017-0177-2. Biol Direct. 2017. PMID: 28187792 Free PMC article. Review. - RNA-guided RNA silencing by an Asgard archaeal Argonaute.
Bastiaanssen C, Bobadilla Ugarte P, Kim K, Finocchio G, Feng Y, Anzelon TA, Köstlbacher S, Tamarit D, Ettema TJG, Jinek M, MacRae IJ, Joo C, Swarts DC, Wu F. Bastiaanssen C, et al. Nat Commun. 2024 Jun 29;15(1):5499. doi: 10.1038/s41467-024-49452-1. Nat Commun. 2024. PMID: 38951509 Free PMC article. - DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins.
Lisitskaya L, Aravin AA, Kulbachinskiy A. Lisitskaya L, et al. Nat Commun. 2018 Dec 4;9(1):5165. doi: 10.1038/s41467-018-07449-7. Nat Commun. 2018. PMID: 30514832 Free PMC article. Review. - Prokaryotic Argonaute Proteins as a Tool for Biotechnology.
Kropocheva EV, Lisitskaya LA, Agapov AA, Musabirov AA, Kulbachinskiy AV, Esyunina DM. Kropocheva EV, et al. Mol Biol. 2022;56(6):854-873. doi: 10.1134/S0026893322060103. Epub 2022 Aug 30. Mol Biol. 2022. PMID: 36060308 Free PMC article.
References
MeSH terms
Substances
Grants and funding
S. W. is supported by the Graduate School for Computing in Medicine and Life Sciences at the University of Lübeck funded by Germany’s Excellence Initiative (DFG GSC 235/1). D. G. acknowledges funding by the Deutsche Forschungsgemeinschaft (GR 3840/2-1). D.G. and T.R. were supported by the Universities of Regensburg and Luebeck, respectively.
LinkOut - more resources
Full Text Sources
Other Literature Sources