Mutant p53 partners in crime - PubMed (original) (raw)
Review
Mutant p53 partners in crime
Michael P Kim et al. Cell Death Differ. 2018 Jan.
Abstract
Mutant p53 proteins impart changes in cellular behavior and function through interactions with proteins that alter gene expression. The milieu of intracellular proteins available to interact with mutant p53 is context specific and changes with disease, cell type, and environmental conditions. Varying conformations of mutant p53 largely dictate protein-protein interactions as different point mutations within protein-coding regions greatly alter the extent and array of gain-of-function (GOF) activities. Given such variables, how can knowledge regarding p53 missense mutations be translated into predicting or altering biologic activity for therapy? How may knowledge regarding mutant p53 functions within certain disease contexts be harnessed to blunt or ablate mutant p53 GOF for therapy? In this article, we review known proteins that interact with mutant p53 and result in the activation of genes that contribute to p53 GOF with particular emphasis on context dependency and an evolving appreciation of GOF mechanisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
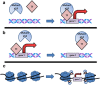
Multiple mechanisms of mutant p53 GOF. (a) Mutant p53 binds to transcription factors (TF) and enhances the transactivation of target genes. (b) Mutant p53 binds various transcription factors and subverts their binding to DNA motifs at the promoters of target genes, leading to reduced expression of target genes. (c) Mutant p53 (light blue circle) modifies the architecture of chromatin by binding and activation of chromatin-modifying enzymes (orange triangle), resulting in enhanced gene expression within spans of accessible chromatin. In this mechanism of mutant p53 GOF, genes occupying entire regions of chromatin may be exposed to existing transcriptional machinery, resulting in gene expression in which gene promoters are not directly bound by mutant p53. Such effects of mutant p53 would not be evident in assays that detect the occupancy of mutant p53 within gene promoters such as ChIP-seq
Figure 2
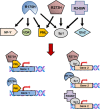
Different mutant p53 proteins converge on common target genes. Multiple mutant p53 proteins (R175H, R273H, R248W) bind to common transcription factors, leading to the activation of identical target genes. For example, mutant p53R175H and R273H both bind the transcription factor PML and transactivate the same target gene (Gene 1). In parallel, mutant p53R273H, R175H, and R248W may bind Sp1 and transactivate an identical target gene (Gene 2). Functionally, different mutant p53 missense mutants (p53R273H, R175H, and R248W) may act through common transcription factors to exert similar GOF through convergence on common target genes. Mutant p53 GOF may therefore depend less on the specific missense mutation site and more on the ability of mutant p53 proteins to bind to core sets of transcription factors and affect their activity
Figure 3
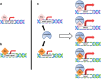
The scope of mutant p53 GOF is enhanced through interactions with multiple transcription factors. Transcription factor 1 (TF 1) and transcription factor 2 (TF2) normally bind to their respective motifs within promoter regions of target genes to initiate transactivation (a, b). Mutant p53R175H may bind to TF1 and TF2 and enhance the expression of target genes specific to each transcription factor. Collectively, through partnerships formed between mutant p53 missense proteins and multiple transcription factors, the influence of mutant p53 on the expression of diverse target genes is amplified. In this scenario, one mutant p53R175H protein may transactivate a total of five target genes through interactions with two different transcription factors (TF1, TF2)
References
- Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 2004; 23: 2330–2338. - PubMed
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004; 119: 861–872. - PubMed
- Milner J, Medcalf EA. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 1991; 65: 765–774. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous