miRNA profiling of NurOwn®: mesenchymal stem cells secreting neurotrophic factors - PubMed (original) (raw)
miRNA profiling of NurOwn®: mesenchymal stem cells secreting neurotrophic factors
Yael Gothelf et al. Stem Cell Res Ther. 2017.
Abstract
Background: MSC-NTF cells are Mesenchymal Stromal Cells (MSC) induced to express high levels of neurotrophic factors (NTFs) using a culture-medium based approach. MSC-NTF cells have been successfully studied in clinical trials for Amyotrophic Lateral Sclerosis (ALS) patients. MicroRNAs (miRNA) are short non-coding RNA molecules that coordinate post-transcriptional regulation of multiple gene targets. The purpose of this study was to determine whether the miRNA profile could provide a tool for MSC-NTF cell characterization and to distinguish them from the matched MSC from which they are derived.
Methods: NTF secretion in the culture supernatant of MSC-NTF cells was evaluated by ELISA assays. The Agilent microarray miRNA platform was used for pairwise comparisons of MSC-NTF cells to MSC. The differentially expressed miRNAs and putative mRNA targets were validated using qPCR analyses.
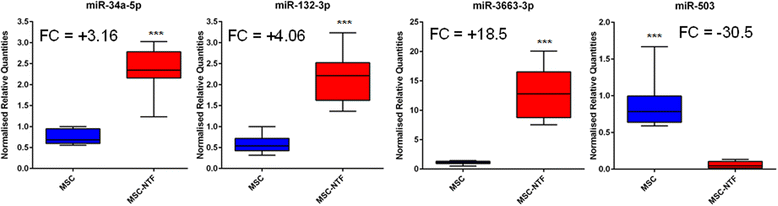
Results: Principal component analysis revealed two distinct clusters based on cell type (MSC and MSC-NTFs). Nineteen miRNAs were found to be upregulated and 22 miRNAs were downregulated in MSC-NTF cells relative to the MSC cells of origin. Further validation of differentially expressed miRNAs confirmed that miR-3663 and miR-132 were increased 18.5- and 4.06-fold, respectively while hsa-miR-503 was reduced more than 15-fold, suggesting that miRNAs could form the basis of an MSC-NTF cell characterization assay. In an analysis of the miRNA mRNA targets, three mRNA targets of hsa-miR-132-3p (HN-1, RASA1 and KLH-L11) were found to be significantly downregulated.
Conclusions: We have demonstrated that MSC-NTF cells can be distinguished from their MSCs of origin by a unique miRNA expression profile.
Trial registration: Clinicaltrial.gov identifier NCT01777646 . Registered 12 December 2012.
Keywords: Amyotrophic Lateral Sclerosis; Mesenchymal Stromal Cells; MicroRNAs; Neurotrophic Factors.
Conflict of interest statement
Authors’ information
Not applicable.
Ethics approval and consent to participate
ALS patients were consented in accordance with the Helsinki declaration in the context of the phase 2a clinical trial
Consent for publication
Not applicable.
Competing interests
Authors are Brainstorm Cell Therapeutics employees
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
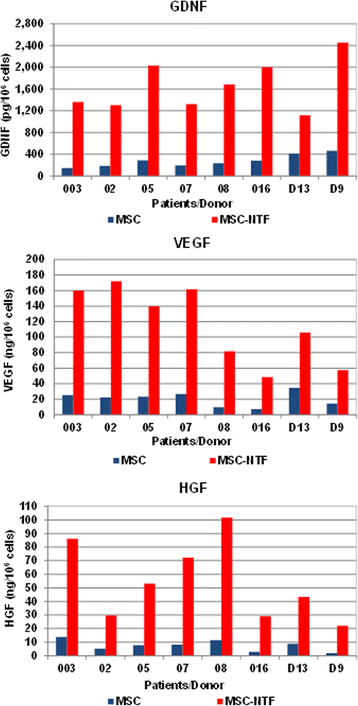
NTF secretion by MSC and MSC-NTF cells of the same patient/donor. MSC of six ALS patients and two healthy donors (D13 and D9) were induced to differentiate into MSC-NTF cells and secretion of neurotrophic factors GDNF, VEGF and HGF was measured in the culture supernatant by ELISA before and after differentiation. Average fold change MSC-NTF/MSC 6.56, 6.01, and 7.85 respectively (p < 0.005). GDNF glial-derived neurotrophic factor, HGF hepatocyte growth factor, MSC mesenchymal stromal cells, NTF neurotrophic factors, VEGF vascular endothelial growth factor
Fig. 2
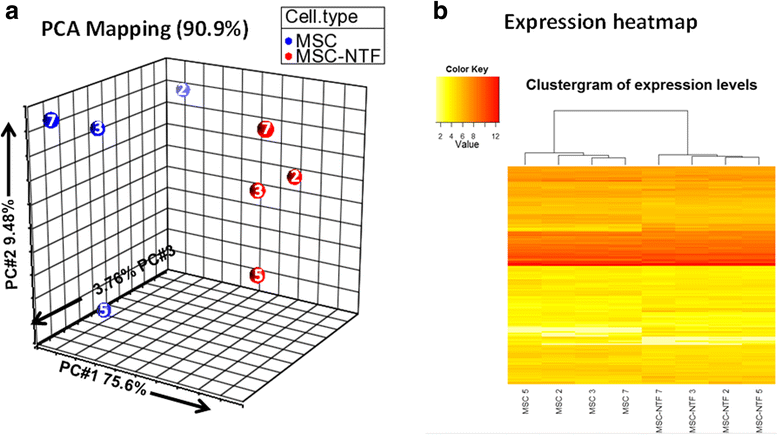
Comparison analysis of the MSC and MSC-NTF cell types based on all 160 detected miRNAs with cell type and donor ID indicated. a Representation of the eight cellular matched miRNA profiles of the four ALS patients in a 3D PCA projection, including donor ID (02, 03, 05, and 07); b Representation of the eight cellular miRNA profiles as a heatmap clustergram plot after hierarchical clustering, including donor ID. MSC mesenchymal stromal cells, NTF neurotrophic factors, PCA principal component analysis
Fig. 3
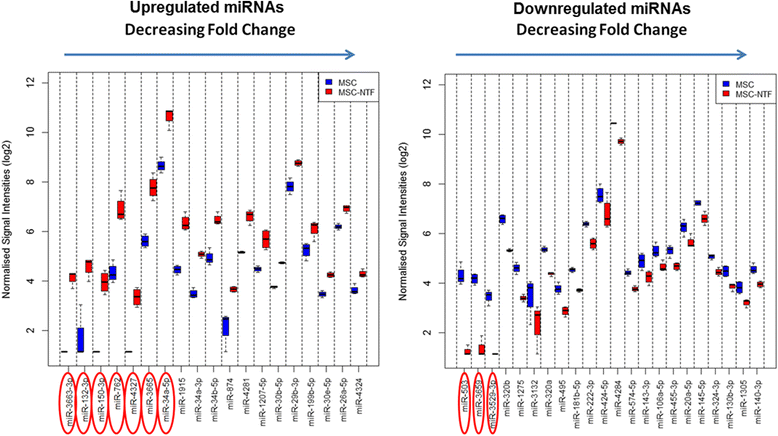
Differentially expressed miRNAs. Expression profiles of the 19 upregulated miRNAs (left panel) and the 22 downregulated miRNAs in MSC-NTF versus MSC on a log2 scale (right panel). The miRNAs most strongly down- and up-regulated in MSC-NTFs are highlighted with red ovals. When the expression of a miRNA was below the level of detection for the arrays, a nominal intensity value is given to these data points to avoid errors arising from non-computable mathematical operations during subsequent data analyses. MSC mesenchymal stromal cells, NTF neurotrophic factors
Fig. 4
Validated differentially expressed miRNAs. Differential expression of miRNAs identified in the microarray was validated by qPCR analysis of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors). ***p < 0.001, two-sided t test. FC fold change, MSC mesenchymal stromal cells, NTF neurotrophic factors
Fig. 5
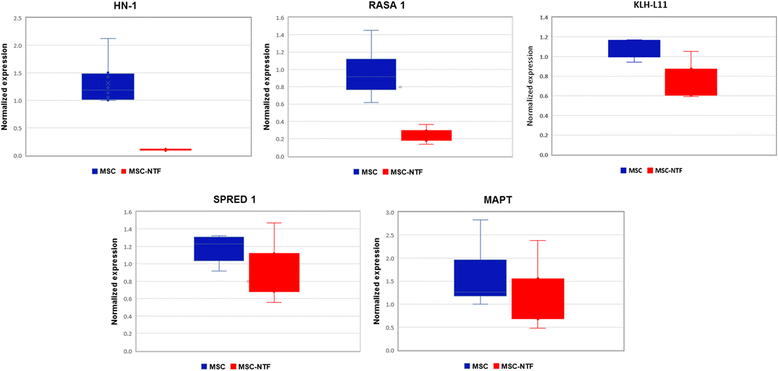
Differential expression of mRNAs targets of hsa-miR-132-3p. Expression of mRNA targets of miR-132-3p were compared in MSC and MSC-NTF cells of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors) by qPCR analysis. HN-1, RASA1 and KLH-L11 were significantly downregulated (11.86 fold, 3.73 -fold and 1.5-fold respectively, p < 0.05). MSC mesenchymal stromal cells, NTF neurotrophic factors
Similar articles
- Safety and Clinical Effects of Mesenchymal Stem Cells Secreting Neurotrophic Factor Transplantation in Patients With Amyotrophic Lateral Sclerosis: Results of Phase 1/2 and 2a Clinical Trials.
Petrou P, Gothelf Y, Argov Z, Gotkine M, Levy YS, Kassis I, Vaknin-Dembinsky A, Ben-Hur T, Offen D, Abramsky O, Melamed E, Karussis D. Petrou P, et al. JAMA Neurol. 2016 Mar;73(3):337-44. doi: 10.1001/jamaneurol.2015.4321. JAMA Neurol. 2016. PMID: 26751635 Clinical Trial. - MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow.
Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B. Gao J, et al. J Cell Biochem. 2011 Jul;112(7):1844-56. doi: 10.1002/jcb.23106. J Cell Biochem. 2011. PMID: 21416501 - MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31.
Baglìo SR, Devescovi V, Granchi D, Baldini N. Baglìo SR, et al. Gene. 2013 Sep 15;527(1):321-31. doi: 10.1016/j.gene.2013.06.021. Epub 2013 Jul 1. Gene. 2013. PMID: 23827457 - Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity.
Shi C, Huang F, Gu X, Zhang M, Wen J, Wang X, You L, Cui X, Ji C, Guo X. Shi C, et al. Oncotarget. 2016 Jun 28;7(26):40830-40845. doi: 10.18632/oncotarget.8518. Oncotarget. 2016. PMID: 27049726 Free PMC article. Review. - [Research progress of mesenchymal stem cell-derived microvesicle].
Wang XQ, Zhu XJ, Zou P. Wang XQ, et al. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013 Feb;21(1):227-30. doi: 10.7534/j.issn.1009-2137.2013.01.046. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013. PMID: 23484725 Review. Chinese.
Cited by
- MicroRNAs in the Migration of Mesenchymal Stem Cells.
He L, Zhang H. He L, et al. Stem Cell Rev Rep. 2019 Feb;15(1):3-12. doi: 10.1007/s12015-018-9852-7. Stem Cell Rev Rep. 2019. PMID: 30328004 Review. - Interaction Between Mesenchymal Stem Cells and Retinal Degenerative Microenvironment.
Lin Y, Ren X, Chen Y, Chen D. Lin Y, et al. Front Neurosci. 2021 Jan 21;14:617377. doi: 10.3389/fnins.2020.617377. eCollection 2020. Front Neurosci. 2021. PMID: 33551729 Free PMC article. Review. - Evaluation of neurotrophic factor secreting mesenchymal stem cells in progressive multiple sclerosis.
Cohen JA, Lublin FD, Lock C, Pelletier D, Chitnis T, Mehra M, Gothelf Y, Aricha R, Lindborg S, Lebovits C, Levy Y, Motamed Khorasani A, Kern R. Cohen JA, et al. Mult Scler. 2023 Jan;29(1):92-106. doi: 10.1177/13524585221122156. Epub 2022 Sep 14. Mult Scler. 2023. PMID: 36113170 Free PMC article. Clinical Trial. - Regulatory effects of miR-28 on osteogenic differentiation of human bone marrow mesenchymal stem cells.
Wang M, Dai T, Meng Q, Wang W, Li S. Wang M, et al. Bioengineered. 2022 Jan;13(1):684-696. doi: 10.1080/21655979.2021.2012618. Bioengineered. 2022. PMID: 34978269 Free PMC article. - Research Trends in the Efficacy of Stem Cell Therapy for Hepatic Diseases Based on MicroRNA Profiling.
Kweon M, Kim JY, Jun JH, Kim GJ. Kweon M, et al. Int J Mol Sci. 2020 Dec 29;22(1):239. doi: 10.3390/ijms22010239. Int J Mol Sci. 2020. PMID: 33383629 Free PMC article. Review.
References
- Petrou P, Gothelf Y, Argov Z, et al. Safety and clinical effects of mesenchymal stem cells secreting neurotrophic factor transplantation in patients with amyotrophic lateral sclerosis: results of phase 1/2 and 2a clinical trials. JAMA Neurol. 2016;73:337–44. doi: 10.1001/jamaneurol.2015.4321. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous