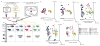Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice - PubMed (original) (raw)
doi: 10.1038/s41564-017-0075-5. Epub 2017 Nov 27.
Thomas Battaglia 3, Yelina Alvarez 3, Luc Bijnens 4, Victoria E Ruiz 3 5, Melody Ho 3, Serina Robinson 6, Tonya Ward 7, Laura M Cox 3 8, Arlin B Rogers 9, Dan Knights 10, R Balfour Sartor 11, Martin J Blaser 12 13 14
Affiliations
- PMID: 29180726
- PMCID: PMC5780248
- DOI: 10.1038/s41564-017-0075-5
Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice
Anjelique F Schulfer et al. Nat Microbiol. 2018 Feb.
Abstract
Antibiotic exposure in children has been associated with the risk of inflammatory bowel disease (IBD). Antibiotic use in children or in their pregnant mother can affect how the intestinal microbiome develops, so we asked whether the transfer of an antibiotic-perturbed microbiota from mothers to their children could affect their risk of developing IBD. Here we demonstrate that germ-free adult pregnant mice inoculated with a gut microbial community shaped by antibiotic exposure transmitted their perturbed microbiota to their offspring with high fidelity. Without any direct or continued exposure to antibiotics, this dysbiotic microbiota in the offspring remained distinct from controls for at least 21 weeks. By using both IL-10-deficient and wild-type mothers, we showed that both inoculum and genotype shape microbiota populations in the offspring. Because IL10-/- mice are genetically susceptible to colitis, we could assess the risk due to maternal transmission of an antibiotic-perturbed microbiota. We found that the IL10-/- offspring that had received the perturbed gut microbiota developed markedly increased colitis. Taken together, our findings indicate that antibiotic exposure shaping the maternal gut microbiota has effects that extend to the offspring, with both ecological and long-term disease consequences.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing financial interests.
Figures
Figure 1. Microbiome transfer to pregnant germ-free mice colonized 23 dams and 112 pups
(a)(i) STAT and Control gut microbiota inocula were taken from a previous experiment.(ii) Germ-free (GF) mice were mated to yield pregnant GF dams. This procedure was done in C57BL/6 mice of wild type (WT) or IL10-deficient (IL10−/−) genotype. (iii) Approximately 1 week before delivery, the pregnant GF dams were gavaged with either the STAT or the Control inoculum, to conventionalize them. Pups were weaned at 4 weeks of age and sacrificed at 21 weeks of age. For the IL10−/− mice, there also were sacrifices at age 6 and 14 weeks. (b) Principal coordinate plots of unweighted UniFrac distances. The inocula used to colonize the dams before delivery are shown together with dam and pup fecal samples from the indicated time points. n=4 replicate aliquots for each inoculum: All numbers of mice/group are shown in Supplementary Table 1, along with P values from Adonis testing.
Figure 2. Intergenerational microbiota transfer efficiency and stability over time
(a) Heat map of relative abundance of OTUs from Control inoculum (C-Inoc), STAT inoculum (S-Inoc), and from dams and pups from all 4 groups of mice at indicated time points (dPG = days post-gavage, wPG = weeks post-gavage). Each row represents an individual mouse microbiota or in the case of the inoculum, a replicate sample. Each column represents an OTU. Phylum level taxonomic assignment of each OTU is indicated by color at the bottom of each column per mouse group. Rows and columns are sorted within each group by OTU prevalence and abundance. n=4 replicates for each inoculum. See Supplementary Table 1 for numbers of mice/group. (b) Average of pairwise Jaccard index values for each pair of samples for each group, as indicated. Pairwise Jaccard distances were generated between pup, dam and inoculum samples for each pup. Summarized heatmaps were created for each group by averaging individuals. (c) Median consecutive pairwise Jaccard distance of dam and pup samples calculated by comparing each animal’s sample to its previous sample, over time from the first time point (1 dPG for dams, 4 wPG for pups) to the last time point (7 wPG for dams, 22 wPG for pups). Boxplots show median values with interquartile range and bars ranging from minimum to maximum values, ** p<0.01, *** p<0.001 one-way ANOVA with Sidak’s multiple comparison test. See Supplementary Table 4 for all P values.
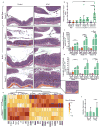
Figure 3. STAT microbiota in IL10−/− mouse increases colonic inflammation
(a) Representative H&E-stained colon sections from WT (top panels, week 21) and IL10−/− mice (bottom 3 rows of panels, weeks 6, 14, and 21) colonized with Control microbiota (left) or STAT microbiota (right). Inset showing inflammation in IL10−/− STAT mice at week 21 at higher magnification on bottom right. Images are representative from a single experiment. Scale bar = 200 μm. (b) Mean histology activity index (HAI) +/− SD for WT and IL10−/− mice colonized with Control or STAT microbiota. ****p<0.0001, Kruskall-Wallis with Dunn’s post-test. (a–b) WT: n = 10 per group; IL10−/−: Control n = 13, 14, 10 and STAT n = 11, 13, 10 at weeks 6, 14, and 21 respectively. (c–d) ELISA results of calprotectin and lipocalin-2 normalized to total protein levels from fecal supernatants. WT: n = 9 per group; IL10−/−: Control n = 10, 11, 10 and STAT n = 9, 10, 9 at weeks 7, 14, and 19 respectively. Mean +/− SEM, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 one way ANOVA with Sidak’s multiple comparison test. (e) Expression of colonic genes that were significantly altered by inoculum in IL10−/− pups (n = 3 each) at 21 weeks, measured by the Nanostring nCounter Mouse Immunology panel. Genes shown in bold and underlined have FDR-adjusted p value < 0.05, by t-test. Genes shown with normal font have FDR-adjusted p value between 0.05 and 0.10. (f) Relative expression of TNF-α and IFN-γ in colonic tissue from week 21 IL10−/− pups, measured by RT-qPCR (STAT TNF-α: n=12; all other groups: n=8), mean +/− SEM *p<0.05 t-test. (a–f) See Supplementary Tables 5–6 for detailed statistics.
Comment in
- Gut microbiota: A mother's microbiota: intergenerational transfer.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2018 Jan;15(1):6. doi: 10.1038/nrgastro.2017.171. Epub 2017 Dec 6. Nat Rev Gastroenterol Hepatol. 2018. PMID: 29209048 No abstract available.
Similar articles
- Early-Life Microbial Restitution Reduces Colitis Risk Promoted by Antibiotic-Induced Gut Dysbiosis in Interleukin 10-/- Mice.
Miyoshi J, Miyoshi S, Delmont TO, Cham C, Lee STM, Sakatani A, Yang K, Shan Y, Kennedy M, Kiefl E, Yousef M, Crosson S, Sogin M, Antonopoulos DA, Eren AM, Leone V, Chang EB. Miyoshi J, et al. Gastroenterology. 2021 Sep;161(3):940-952.e15. doi: 10.1053/j.gastro.2021.05.054. Epub 2021 Jun 7. Gastroenterology. 2021. PMID: 34111469 Free PMC article. - Dietary iron variably modulates assembly of the intestinal microbiota in colitis-resistant and colitis-susceptible mice.
Ellermann M, Gharaibeh RZ, Maharshak N, Peréz-Chanona E, Jobin C, Carroll IM, Arthur JC, Plevy SE, Fodor AA, Brouwer CR, Sartor RB. Ellermann M, et al. Gut Microbes. 2020;11(1):32-50. doi: 10.1080/19490976.2019.1599794. Epub 2019 Jun 10. Gut Microbes. 2020. PMID: 31179826 Free PMC article. - Metagenomic Alterations in Gut Microbiota Precede and Predict Onset of Colitis in the IL10 Gene-Deficient Murine Model.
Miyoshi J, Lee STM, Kennedy M, Puertolas M, Frith M, Koval JC, Miyoshi S, Antonopoulos DA, Leone V, Chang EB. Miyoshi J, et al. Cell Mol Gastroenterol Hepatol. 2021;11(2):491-502. doi: 10.1016/j.jcmgh.2020.08.008. Epub 2020 Aug 21. Cell Mol Gastroenterol Hepatol. 2021. PMID: 32835897 Free PMC article. - Interrelatedness between dysbiosis in the gut microbiota due to immunodeficiency and disease penetrance of colitis.
Ray A, Dittel BN. Ray A, et al. Immunology. 2015 Nov;146(3):359-68. doi: 10.1111/imm.12511. Epub 2015 Sep 7. Immunology. 2015. PMID: 26211540 Free PMC article. Review. - Gut microbiome in primary sclerosing cholangitis: A review.
Little R, Wine E, Kamath BM, Griffiths AM, Ricciuto A. Little R, et al. World J Gastroenterol. 2020 Jun 7;26(21):2768-2780. doi: 10.3748/wjg.v26.i21.2768. World J Gastroenterol. 2020. PMID: 32550753 Free PMC article. Review.
Cited by
- The Microbiome and Infectious Diseases.
Haraoui LP, Blaser MJ. Haraoui LP, et al. Clin Infect Dis. 2023 Dec 5;77(Suppl 6):S441-S446. doi: 10.1093/cid/ciad577. Clin Infect Dis. 2023. PMID: 38051971 Free PMC article. - Metagenomic tracking of antibiotic resistance genes through a pre-harvest vegetable production system: an integrated lab-, microcosm- and greenhouse-scale analysis.
Keenum I, Wind L, Ray P, Guron G, Chen C, Knowlton K, Ponder M, Pruden A. Keenum I, et al. Environ Microbiol. 2022 Aug;24(8):3705-3721. doi: 10.1111/1462-2920.16022. Epub 2022 May 18. Environ Microbiol. 2022. PMID: 35466491 Free PMC article. - Species abundance information improves sequence taxonomy classification accuracy.
Kaehler BD, Bokulich NA, McDonald D, Knight R, Caporaso JG, Huttley GA. Kaehler BD, et al. Nat Commun. 2019 Oct 11;10(1):4643. doi: 10.1038/s41467-019-12669-6. Nat Commun. 2019. PMID: 31604942 Free PMC article. - Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract.
Fenneman AC, Weidner M, Chen LA, Nieuwdorp M, Blaser MJ. Fenneman AC, et al. Nat Rev Gastroenterol Hepatol. 2023 Feb;20(2):81-100. doi: 10.1038/s41575-022-00685-9. Epub 2022 Oct 18. Nat Rev Gastroenterol Hepatol. 2023. PMID: 36258032 Free PMC article. Review. - Gut microbiome of treatment-naïve MS patients of different ethnicities early in disease course.
Ventura RE, Iizumi T, Battaglia T, Liu M, Perez-Perez GI, Herbert J, Blaser MJ. Ventura RE, et al. Sci Rep. 2019 Nov 8;9(1):16396. doi: 10.1038/s41598-019-52894-z. Sci Rep. 2019. PMID: 31705027 Free PMC article.
References
- Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. - PubMed
- Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes. 2014;38:1290–1298. - PubMed
- Arrieta M-C, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152–307ra152. - PubMed
- Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. - PubMed
References for Methods
- Cox D, Snell EJ. Analysis of Binary Data. Chapman and Hall/CRC press; 1989.
- Aronesty E. Comparison of Sequencing Utility Programs. Open Bioinforma J. 2013;7:1–8.
Publication types
MeSH terms
Substances
Grants and funding
- P01 DK094779/DK/NIDDK NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- P40 OD010995/OD/NIH HHS/United States
- P30 DK034987/DK/NIDDK NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R01 DK090989/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical