Using deep learning to model the hierarchical structure and function of a cell - PubMed (original) (raw)
Using deep learning to model the hierarchical structure and function of a cell
Jianzhu Ma et al. Nat Methods. 2018 Apr.
Abstract
Although artificial neural networks are powerful classifiers, their internal structures are hard to interpret. In the life sciences, extensive knowledge of cell biology provides an opportunity to design visible neural networks (VNNs) that couple the model's inner workings to those of real systems. Here we develop DCell, a VNN embedded in the hierarchical structure of 2,526 subsystems comprising a eukaryotic cell (http://d-cell.ucsd.edu/). Trained on several million genotypes, DCell simulates cellular growth nearly as accurately as laboratory observations. During simulation, genotypes induce patterns of subsystem activities, enabling in silico investigations of the molecular mechanisms underlying genotype-phenotype associations. These mechanisms can be validated, and many are unexpected; some are governed by Boolean logic. Cumulatively, 80% of the importance for growth prediction is captured by 484 subsystems (21%), reflecting the emergence of a complex phenotype. DCell provides a foundation for decoding the genetics of disease, drug resistance and synthetic life.
Conflict of interest statement
Competing financial interests
Trey Ideker is co-founder of Data4Cure, Inc. and has an equity interest. Trey Ideker has an equity interest in Ideaya BioSciences, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Figures
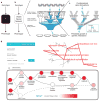
Figure 1. Modeling system structure and function with visible learning
a, A conventional neural network translates input to output as a black box without knowledge of system structure. b, In a visible neural network, input/output translation is based on prior structural knowledge. In DCell, gene disruption genotypes (top) are translated to cell growth predictions (bottom) through a hierarchy of cell subsystems (middle). c, A neural network is embedded in the prior structure using multiple neurons per subsystem. d, Screen capture of DCell online service, microtubule subsystems.
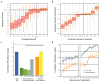
Figure 2. Prediction of cell viability and genetic interaction phenotypes
a, Measured versus predicted cell viability relative to wild type (WT = 1) on the Costanzo et al. dataset. b, Measured versus predicted genetic interaction scores for each double gene disruption genotype; genetic interactions between the disrupted genes can be positive (epistasis), zero (non-interaction), or negative (synthetic sickness or lethality). c, Model performance expressed as the correlation between measured and predicted genetic interaction scores. Performance of DCell (first bar, blue) is compared to previous methods for predicting genetic interactions (second four bars, shades of green): FBA, Flux Balance Analysis; GBA, Guilt By Association; MNMC, Multi-Network Multi-Classifier; Ontotype. Performance is also shown for matched black-box structures in which gene-to-subsystem mappings are randomly permuted (orange bar, average of 10 randomizations) or for fully-connected neural networks with the same number of layers as DCell (final bar, yellow). Correlations were calculated across gene pairs that meet an interaction significance criterion of p<0.05. d, Predictive performance versus number of neurons per subsystem. Performance measure and two structural hierarchies as in (c).
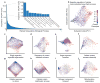
Figure 3. Interpretation of genotype-phenotype associations
a, Hierarchy of candidate subsystems that can explain the association of the pmt1Δire1Δ genotype with a negative genetic interaction phenotype (synthetic lethality). Subsystem states are represented by their neuron values, reduced to the first two principal components (PCs) which captured >75% of the total variance. The color of the node for each subsystem shows its PC2 expressed as a z-score with zero mean and unit standard deviation. By construction, the wild-type genotype produces z-score = 0 for all subsystems (Online Methods). b, Correspondence between Hac1 GFP activity and the functional states of ER-UPR (blue) or its parent subsystem, Response to ER stress (red). Points represent genotypes, with pmt1Δire1Δ genotype indicated. Subsystem state is the z-score of PC2 as in (a). c, Distribution of correlations between Hac1 GFP activity and the states of every other subsystem containing at least 10 genotypes with measured GFP activity. d, DNA repair is a highly altered subsystem that explains the slow growth phenotype of rev7Δrad57Δ. e, Experimental resistance to UV damage plotted against the simulated state of DNA repair in DCell, separated into two classes: above or below wild-type value. Significance measured by Mann–Whitney U test. f, Distribution of the associations between UV damage resistance and the states of other subsystems containing at least 10 genotypes with measured UV resistance. Panels (a–c) use genetic interaction prediction as the phenotypic readout; (d–f) use growth. All panels implement GO as the structural model.
Figure 4. Top subsystem states for translation of genotype to growth
a, Ranking of all cellular subsystems in GO (x-axis) by their importance in determining genetic interactions (RLIPP score, y-axis). Inset: ten highest-scoring subsystems. Positive RLIPP corresponds to increases in predictive power relative to children; negative RLIPP corresponds to decreases in power. b–j, Two-dimensional state maps of informative subsystems (PC2 versus PC1). Supplementary Fig. 2 provides equivalent information for the CliXO hierarchy.
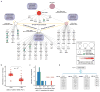
Figure 5. Analysis of subsystem functional logic
a, Causal hierarchy of important subsystems involving changes to Mitochondrial Respiratory Chain (MRC), displayed as in Fig. 3a. b, 2D state map of MRC plotted as in Fig. 3b. Genotypes disrupting MRC complex III only, MRC complex IV only, or both complexes are demarcated by colors. c, Truth table relating the state of MRC to the states of its children. The logic resembles an AND gate, pictured. d, Newly identified system (CliXO:10651) encoding a logical AND integrating the states of two well-characterized children. Names of children are cross-annotated from the corresponding enriched GO terms: Actin filament-based process, GO:0030029; Monovalent inorganic ion homeostasis, GO:0030004.
Figure 6. Analysis of a new DNA repair subsystem
a, Hierarchical organization of DNA repair including subsystems newly identified by CliXO. b, Experimental resistance to UV damage plotted against the state of CliXO:10582, separated into two classes: above or below wild-type value. Significance measured by Mann–Whitney U test. c, Distribution of associations between UV resistance and the states of all CliXO subsystems with at least 10 genotypes with measured UV resistance. d, Approximate mathematical function implemented by CliXO:10582 neurons.
Comment in
- A deep (learning) dive into a cell.
Branson K. Branson K. Nat Methods. 2018 Apr 3;15(4):253-254. doi: 10.1038/nmeth.4658. Nat Methods. 2018. PMID: 29614064 No abstract available.
Similar articles
- Prediction of anticancer drug sensitivity using an interpretable model guided by deep learning.
Pang W, Chen M, Qin Y. Pang W, et al. BMC Bioinformatics. 2024 May 9;25(1):182. doi: 10.1186/s12859-024-05669-x. BMC Bioinformatics. 2024. PMID: 38724920 Free PMC article. - A structure-time parallel implementation of spike-based deep learning.
Wu X, Wang Y, Tang H, Yan R. Wu X, et al. Neural Netw. 2019 May;113:72-78. doi: 10.1016/j.neunet.2019.01.010. Epub 2019 Feb 4. Neural Netw. 2019. PMID: 30785011 - Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model.
Chen L, Cai C, Chen V, Lu X. Chen L, et al. BMC Bioinformatics. 2016 Jan 11;17 Suppl 1(Suppl 1):9. doi: 10.1186/s12859-015-0852-1. BMC Bioinformatics. 2016. PMID: 26818848 Free PMC article. - From chemoinformatics to deep learning: an open road to drug discovery.
Ferreira LL, Andricopulo AD. Ferreira LL, et al. Future Med Chem. 2019 Mar;11(5):371-374. doi: 10.4155/fmc-2018-0449. Epub 2019 Mar 19. Future Med Chem. 2019. PMID: 30888845 Review. No abstract available. - Deep Learning in Drug Discovery.
Gawehn E, Hiss JA, Schneider G. Gawehn E, et al. Mol Inform. 2016 Jan;35(1):3-14. doi: 10.1002/minf.201501008. Epub 2015 Dec 30. Mol Inform. 2016. PMID: 27491648 Review.
Cited by
- pSuc-EDBAM: Predicting lysine succinylation sites in proteins based on ensemble dense blocks and an attention module.
Jia J, Wu G, Li M, Qiu W. Jia J, et al. BMC Bioinformatics. 2022 Oct 31;23(1):450. doi: 10.1186/s12859-022-05001-5. BMC Bioinformatics. 2022. PMID: 36316638 Free PMC article. - Semantic similarity and machine learning with ontologies.
Kulmanov M, Smaili FZ, Gao X, Hoehndorf R. Kulmanov M, et al. Brief Bioinform. 2021 Jul 20;22(4):bbaa199. doi: 10.1093/bib/bbaa199. Brief Bioinform. 2021. PMID: 33049044 Free PMC article. Review. - Computer-Aided Whole-Cell Design: Taking a Holistic Approach by Integrating Synthetic With Systems Biology.
Marucci L, Barberis M, Karr J, Ray O, Race PR, de Souza Andrade M, Grierson C, Hoffmann SA, Landon S, Rech E, Rees-Garbutt J, Seabrook R, Shaw W, Woods C. Marucci L, et al. Front Bioeng Biotechnol. 2020 Aug 7;8:942. doi: 10.3389/fbioe.2020.00942. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32850764 Free PMC article. - Multimodal learning with graphs.
Ektefaie Y, Dasoulas G, Noori A, Farhat M, Zitnik M. Ektefaie Y, et al. Nat Mach Intell. 2023 Apr;5(4):340-350. doi: 10.1038/s42256-023-00624-6. Epub 2023 Apr 3. Nat Mach Intell. 2023. PMID: 38076673 Free PMC article. - The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions.
Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, Dolma S, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. Oughtred R, et al. Protein Sci. 2021 Jan;30(1):187-200. doi: 10.1002/pro.3978. Epub 2020 Nov 23. Protein Sci. 2021. PMID: 33070389 Free PMC article.
References
- Farabet C, Couprie C, Najman L, Lecun Y. Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Mach Intell. 2013;35:1915–1929. - PubMed
- Mikolov T, Deoras A, Povey D, Burget L, Černocký J. Strategies for training large scale neural network language models. in. 2011 IEEE Workshop on Automatic Speech Recognition Understanding; 2011. pp. 196–201.
- Hinton G, et al. Deep Neural Networks for Acoustic Modeling in Speech Recognition: The Shared Views of Four Research Groups. IEEE Signal Process Mag. 2012;29:82–97.
- Sainath TN, Mohamed Ar, Kingsbury B, Ramabhadran B. Deep convolutional neural networks for LVCSR. 2013 IEEE International Conference on Acoustics, Speech and Signal Processing; 2013. pp. 8614–8618.
- Collobert R, et al. Natural Language Processing (Almost) from Scratch. J Mach Learn Res. 2011;12:2493–2537.
Publication types
MeSH terms
Grants and funding
- OT3 TR002026/TR/NCATS NIH HHS/United States
- P41 GM103504/GM/NIGMS NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
- R01 HG009979/HG/NHGRI NIH HHS/United States
- U54 CA209891/CA/NCI NIH HHS/United States
- R01 GM084279/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources