Redefining the ancestral origins of the interleukin-1 superfamily - PubMed (original) (raw)
Redefining the ancestral origins of the interleukin-1 superfamily
Jack Rivers-Auty et al. Nat Commun. 2018.
Abstract
The interleukin-1 (IL-1) receptor and ligand families are components of the immune system. Knowledge of their evolutionary history is essential to understand their function. Using chromosomal anatomy and sequence similarity, we show that IL-1 receptor family members are related and nine members are likely formed from duplication and modification of a proto-IL-1R1 receptor. The IL-1 ligands have a different evolutionary history. The first proto-IL-1β gene coincided with proto-IL-1R1 and duplication events resulted in the majority of IL-1 ligand family members. However, large evolutionary distances are observed for IL-1α, IL-18 and IL-33 proteins. Further analysis show that IL-33 and IL-18 have poor sequence similarity and no chromosomal evidence of common ancestry with the IL-1β cluster and therefore should not be included in the IL-1 ligand ancestral family. IL-1α formed from the duplication of IL-1β, and moonlighting functions of pro-IL-1α acted as divergent selection pressures for the observed sequence dissimilarity.
Conflict of interest statement
The authors declare no competing interests.
Figures
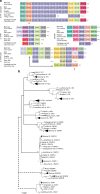
Fig. 1
Evolutionary history of the IL-1 receptor family. a Chromosomal gene location as evidence of ancestral relationship of IL-1 family members. The highly conserved nature of surrounding genes and sequence homology suggests two or one separate evolutionary families: IL-1R1, IL-1R2, IL-18R1, IL-1RL2, IL-1RL2 and IL-18RAP all form a cluster on the same chromosome, indicating gene duplication events. IL-1RAPL2 also has the MAP4K4 gene nearby indicating a duplication and translocation of that part of the chromosome. The ARX gene indicates that IL-1RAPL1 likely formed from a duplication of IL-1RAPL2. IL-1RAP shares no nearby genes but demonstrates high sequence identity with the IL-18RAP gene inferring homology. This suggests there is clear evidence that all the IL-1R superfamily genes have a common ancestor except for the SIGIRR gene, which has relatively low sequence identity and no chromosomal anatomy evidence to support shared ancestry. b Composite evolutionary history of the IL-1 family of cytokines constructed by overlaying the evidence from chromosomal location and clade IL-1 family gene profile on to the maximum likelihood tree from Supplementary Data 1. Dotted lines represent less established evidence of shared ancestry. The percentage of trees in which the associated group clustered together is shown next to the branches from 1000 bootstrap replications. Branches, which occur in <50% of trees were collapsed. The tree is drawn to scale, with branch lengths measured in the number of amino acid replacements per site. The analysis involved 231 amino acid sequences with an alignment length of 64 positions in the final data set. All positions containing gaps and missing data were eliminated. Scale bar is 0.5 replacements per site
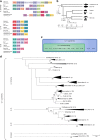
Fig. 2
Evolutionary history of the IL-1 ligand family. a Chromosomal gene location as evidence of ancestral relationship of IL-1 family members. The highly conserved nature of surrounding genes suggests three separate evolutionary families, as follows: IL-1β, IL-18, and IL-33. b A simplified evolutionary tree with time scale of cartilaginous and bony fish, birds, reptiles, mammals and the approximate time line of the evolutionary occurrence of IL-1 family members,. IL-1β and IL-18 are expressed exclusively in all vertebrate species, including cartilaginous fish, suggesting that they evolved prior to the divergence of bony and cartilaginous fish ~425 million years ago (ii); IL-1α, IL-33 and IL-36 α, β & γ are expressed exclusively in mammals therefore likely formed in the common ancestor of all mammals (Synapsid lineage) (iv). This event must have occurred after divergence of the Synapsid lineage (iv) 320 million years ago but prior to the divergence of mammals 160 million years ago (v). c An ancestral and superfamily scheme of the IL-1 ligands. d Composite evolutionary history of the IL-1 family of cytokines constructed by overlaying the evidence from chromosomal location and clade IL-1 family gene profile on to the maximum likelihood tree from Supplementary Data 2. The percentage of trees in which the associated group clustered together is shown next to the branches from 1000 bootstrap replications. Primary branches that occur in <50% of trees were collapsed. The tree is drawn to scale, with branch lengths measured in the number of amino acid replacements per site. The analysis involved 155 amino acid sequences with a final alignment length of 64 positions. All positions containing gaps and missing data were eliminated. Scale bar is 0.5 replacements per site
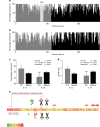
Fig. 3
Conservation of IL-1α and IL-1β sequences in mammals. The percentage of conserved a.a. residues of the pro (grey) and mature (black) segments of IL-1α (a) and IL-1β (b). c The average conservation from a, b showing that the amino acid sequence of the pro-domain of IL-1α is highly conserved, more so than the mature domain of either IL-1α or IL-1β. d The ratio of synonymous (dS) to non-synonymous (dN) substitutions per potential substitution site demonstrates strong positive selection for the conserved pro-domain of IL-1α. Analyses were conducted using the Nei-Gojobori method. e A heat map of the conservation of IL-1α sequence across mammalian species depicting the correlation with putative functions of the IL-1α protein (green 25% conserved to red 100% conserved across mammalian species). The NLS sequence, histone acetyltransferase complex-binding region, HAX-1-binding region, neutrophil elastase cleavage site (N) and NLS post-translation modification sites were very highly conserved (P, phosphorylation; A, acetylation; M, myristoylation), while the glycosylation (gly) site, RNA splicing domain and cleavage sites for chymase (Ch), caspase-1 (C), and granzyme B (G) were poorly conserved. c, d bars are means ± SEM, *p < 0.05 and **p < 0.01 pro-domain vs mature domain within IL-1 member; #p < 0.05 and ##p < 0.01 IL-1β vs. IL-1α within domain, omnibus effects evaluated by linear modelling followed Sidak correct post-hoc tests
Fig. 4
The evolutionary importance of HAX-1 to IL-1α. a A model evolutionary tree with time scale of cartilaginous and body fish, birds, reptiles, mammals and baleen and toothed whales,. HAX-1 is expressed in all vertebrate species as well as hemichordates, suggesting that it emerged during the Cambrian explosion (i); IL-1β evolved approximately 425 million years ago (ii); IL-1α formed via a gene duplication event of IL-1β in the common ancestor of all mammals (Synapsid lineage) (iv). The NLS of IL-1α is present in all land mammals and baleen whales, suggesting it was present in the common ancestor of all mammals (v) and the common ancestor of all whales (vi); all toothed whales lack a functional NLS, suggesting that the common ancestor of the toothed whale (vii) had a loss of function mutation in the NLS. b The amino acid sequences of the IL-1α NLS, including the percentage conservation and the calculated predicted NLS activity (cNLS) scores; all found in the pro-region in amino acid positions around 70–85. Note the loss of a functional NLS in the toothed whale species due in most part to the R to W replacement (highlighted). c The percentage of conserved amino acid sequences of the pro (grey) and mature (black) segments of IL-1α comparing 4 toothed whale species with the modal land mammal sequence. While the mature segment contains substantial variation due to the build-up of neutral mutations over the 35 million years of evolutionary separation, the pro-IL-1α domain is highly conserved despite the loss of NLS function. d Dot blot with bovine serum albumin (BSA) (1), pro-IL-1α (2), mature IL-1α (3), mature IL-1β (4) and HAX-1 (5) bound to the membrane (0.02 μg), then incubated with HAX-1 (5 μg ml−1) (HAX) or control buffer (Con) to investigate HAX-1 binding. HAX-1 was probed and visualised, and found to exclusively bind to pro-IL-1α, confirming that this binding is specific and substantial to the pro-domain of pro-IL-1α
Similar articles
- Interleukin-1 Ligands and Receptors in Lumpfish (Cyclopterus lumpus L.): Molecular Characterization, Phylogeny, Gene Expression, and Transcriptome Analyses.
Eggestøl HØ, Lunde HS, Knutsen TM, Haugland GT. Eggestøl HØ, et al. Front Immunol. 2020 Apr 2;11:502. doi: 10.3389/fimmu.2020.00502. eCollection 2020. Front Immunol. 2020. PMID: 32300342 Free PMC article. - Altered interleukin-1 receptor antagonist and interleukin-18 mRNA expression in myocardial tissues of patients with dilatated cardiomyopathy.
Westphal E, Rohrbach S, Buerke M, Behr H, Darmer D, Silber RE, Werdan K, Loppnow H. Westphal E, et al. Mol Med. 2008 Jan-Feb;14(1-2):55-63. doi: 10.2119/2007-00058.Westphal. Mol Med. 2008. PMID: 17948066 Free PMC article. - Postoperative ileus involves interleukin-1 receptor signaling in enteric glia.
Stoffels B, Hupa KJ, Snoek SA, van Bree S, Stein K, Schwandt T, Vilz TO, Lysson M, Veer CV, Kummer MP, Hornung V, Kalff JC, de Jonge WJ, Wehner S. Stoffels B, et al. Gastroenterology. 2014 Jan;146(1):176-87.e1. doi: 10.1053/j.gastro.2013.09.030. Epub 2013 Sep 22. Gastroenterology. 2014. PMID: 24067878 - The molecular evolution of the interleukin-1 family of cytokines; IL-18 in teleost fish.
Huising MO, Stet RJ, Savelkoul HF, Verburg-van Kemenade BM. Huising MO, et al. Dev Comp Immunol. 2004 May 3;28(5):395-413. doi: 10.1016/j.dci.2003.09.005. Dev Comp Immunol. 2004. PMID: 15062640 Review. - The modern interleukin-1 superfamily: Divergent roles in obesity.
Lee MK, Yvan-Charvet L, Masters SL, Murphy AJ. Lee MK, et al. Semin Immunol. 2016 Oct;28(5):441-449. doi: 10.1016/j.smim.2016.10.001. Epub 2016 Oct 7. Semin Immunol. 2016. PMID: 27726910 Review.
Cited by
- Post-stroke inflammation-target or tool for therapy?
Lambertsen KL, Finsen B, Clausen BH. Lambertsen KL, et al. Acta Neuropathol. 2019 May;137(5):693-714. doi: 10.1007/s00401-018-1930-z. Epub 2018 Nov 27. Acta Neuropathol. 2019. PMID: 30483945 Free PMC article. Review. - The Paradigm Change of IL-33 in Vascular Biology.
Demyanets S, Stojkovic S, Huber K, Wojta J. Demyanets S, et al. Int J Mol Sci. 2021 Dec 10;22(24):13288. doi: 10.3390/ijms222413288. Int J Mol Sci. 2021. PMID: 34948083 Free PMC article. Review. - IL-36 promotes anti-viral immunity by boosting sensitivity to IFN-α/β in IRF1 dependent and independent manners.
Wang P, Gamero AM, Jensen LE. Wang P, et al. Nat Commun. 2019 Oct 16;10(1):4700. doi: 10.1038/s41467-019-12318-y. Nat Commun. 2019. PMID: 31619669 Free PMC article. - Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity.
Mantovani A, Dinarello CA, Molgora M, Garlanda C. Mantovani A, et al. Immunity. 2019 Apr 16;50(4):778-795. doi: 10.1016/j.immuni.2019.03.012. Immunity. 2019. PMID: 30995499 Free PMC article. Review. - Revisiting the origin of interleukin 1 in anamniotes and sub-functionalization of interleukin 1 in amniotes.
Hasel de Carvalho E, Bartok E, Stölting H, Bajoghli B, Leptin M. Hasel de Carvalho E, et al. Open Biol. 2022 Aug;12(8):220049. doi: 10.1098/rsob.220049. Epub 2022 Aug 17. Open Biol. 2022. PMID: 35975650 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous