FusoPortal: an Interactive Repository of Hybrid MinION-Sequenced Fusobacterium Genomes Improves Gene Identification and Characterization - PubMed (original) (raw)
FusoPortal: an Interactive Repository of Hybrid MinION-Sequenced Fusobacterium Genomes Improves Gene Identification and Characterization
Blake E Sanders et al. mSphere. 2018.
Erratum in
- Erratum for Sanders et al., "FusoPortal: an Interactive Repository of Hybrid MinION-Sequenced Fusobacterium Genomes Improves Gene Identification and Characterization".
Sanders BE, Umana A, Lemkul JA, Slade DJ. Sanders BE, et al. mSphere. 2018 Jul 25;3(4):e00379-18. doi: 10.1128/mSphere.00379-18. mSphere. 2018. PMID: 30156798 Free PMC article. No abstract available.
Abstract
Here we present FusoPortal, an interactive repository of Fusobacterium genomes that were sequenced using a hybrid MinION long-read sequencing pipeline, followed by assembly and annotation using a diverse portfolio of predominantly open-source software. Significant efforts were made to provide genomic and bioinformatic data as downloadable files, including raw sequencing reads, genome maps, gene annotations, protein functional analysis and classifications, and a custom BLAST server for FusoPortal genomes. FusoPortal has been initiated with eight complete genomes, of which seven were previously only drafts that ranged from 24 to 67 contigs. We have showcased that the genomes in FusoPortal provide accurate open reading frame annotations and have corrected a number of large (>3-kb) genes that were previously misannotated due to contig boundaries. In summary, FusoPortal (http://fusoportal.org) is the first database of MinION-sequenced and completely assembled Fusobacterium genomes, and this central Fusobacterium genomic and bioinformatic resource will aid the scientific community in developing a deeper understanding of how this human pathogen contributes to an array of diseases, including periodontitis and colorectal cancer.IMPORTANCE In this report, we describe a hybrid MinION whole-genome sequencing pipeline and the genomic characteristics of the first eight Fusobacterium strains deposited in the FusoPortal database. This collection of highly accurate and complete genomes drastically improves upon previous multicontig assemblies by correcting and newly identifying a significant number of open reading frames. We believe that the availability of this resource will result in the discovery of proteins and molecular mechanisms used by an oral pathogen, with the potential to further our understanding of how Fusobacterium nucleatum contributes to a repertoire of diseases, including periodontitis, preterm birth, and colorectal cancer.
Keywords: Fusobacterium; Fusobacterium nucleatum; colon cancer; colorectal cancer; genomics; host-pathogen; pathogenesis.
Copyright © 2018 Sanders et al.
Figures
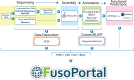
FIG 1
Schematic of all genomic, bioinformatic, and scripting workflows used to create FusoPortal.
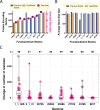
FIG 2
Correction of open reading frames in Fusobacterium genomes. (A) A comparison of the genome size to the number of protein-encoding genes per genome in both the NCBI and FusoPortal genomes. (B) A comparison of all proteins for average gene size in the NCBI and FusoPortal genomes. (C) Analysis of all proteins 1,000 residues in size and above in annotated FusoPortal proteomes and of how many residues were added (or, in rare cases, removed) compared to the previous annotations present in the NCBI database. n = number of proteins 1,000 residues in size or greater in each genome.
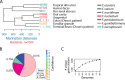
FIG 3
Phylogenetic analysis of complete Fusobacterium genomes. (A) Tree created using complete proteomes in the microPan package in R. Bootstrap values are indicated in red. Locations of where strains were isolated in the human body are highlighted. (B) Micropan analysis of pan-genome gene families with models predicting the percentages of core genes, shell genes (present in most genomes), and cloud genes (found in few genomes). (C) Analysis of the number of unique gene clusters found among the eight complete FusoPortal genomes.
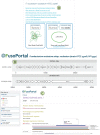
FIG 4
Navigating the FusoPortal Web interface for genome analysis. (A) Links are provided for downloading all genomic data (raw and analyzed), with Bandage plots to show the completeness of each genome build. (B) Whole-genome maps were produced with links to custom webpages for each gene that provide access to all genomic and bioinformatic analysis. (C) Each gene contains a full InterPro analysis page with links to multiple functional prediction databases (e.g., PFAM, TIGR, Gene3D, CDD, GO). No IPR, no InterPro accession number.
FIG 5
A custom BLAST database to search FusoPortal genomes built with SequenceServer. (A) Data entry panel that autodetects protein or DNA sequences. (B) The available genomes that users can choose to search. (C) BLAST results for identified genes. (D) Links to downloadable files.
Similar articles
- Fusobacterium Genomics Using MinION and Illumina Sequencing Enables Genome Completion and Correction.
Todd SM, Settlage RE, Lahmers KK, Slade DJ. Todd SM, et al. mSphere. 2018 Jul 5;3(4):e00269-18. doi: 10.1128/mSphere.00269-18. mSphere. 2018. PMID: 29976645 Free PMC article. - Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article. - Erratum for Sanders et al., "FusoPortal: an Interactive Repository of Hybrid MinION-Sequenced Fusobacterium Genomes Improves Gene Identification and Characterization".
Sanders BE, Umana A, Lemkul JA, Slade DJ. Sanders BE, et al. mSphere. 2018 Jul 25;3(4):e00379-18. doi: 10.1128/mSphere.00379-18. mSphere. 2018. PMID: 30156798 Free PMC article. No abstract available. - Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing.
Goldstein S, Beka L, Graf J, Klassen JL. Goldstein S, et al. BMC Genomics. 2019 Jan 9;20(1):23. doi: 10.1186/s12864-018-5381-7. BMC Genomics. 2019. PMID: 30626323 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
- Fusobacterium Genomics Using MinION and Illumina Sequencing Enables Genome Completion and Correction.
Todd SM, Settlage RE, Lahmers KK, Slade DJ. Todd SM, et al. mSphere. 2018 Jul 5;3(4):e00269-18. doi: 10.1128/mSphere.00269-18. mSphere. 2018. PMID: 29976645 Free PMC article. - Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article. - Colorectal cancer: the facts in the case of the microbiota.
Clay SL, Fonseca-Pereira D, Garrett WS. Clay SL, et al. J Clin Invest. 2022 Feb 15;132(4):e155101. doi: 10.1172/JCI155101. J Clin Invest. 2022. PMID: 35166235 Free PMC article. Review. - Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review. - Fusobacterium nucleatum Metabolically Integrates Commensals and Pathogens in Oral Biofilms.
Sakanaka A, Kuboniwa M, Shimma S, Alghamdi SA, Mayumi S, Lamont RJ, Fukusaki E, Amano A. Sakanaka A, et al. mSystems. 2022 Aug 30;7(4):e0017022. doi: 10.1128/msystems.00170-22. Epub 2022 Jul 19. mSystems. 2022. PMID: 35852319 Free PMC article.
References
- Signat B, Roques C, Poulet P, Duffaut D. 2011. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13:25–36. - PubMed
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi:10.1101/gr.126573.111. - DOI - PMC - PubMed
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi:10.1016/j.chom.2013.07.007. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials