Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string - PubMed (original) (raw)
Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string
Qi Hao et al. Cell Discov. 2018.
No abstract available
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
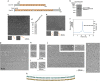
Fig. 1. Electron microscopy of _Ct_pom152 reveals the assembly of beaded strings.
a Domain architecture of _Ct_pom152 and _Hs_gp210. Regions without a predicted fold are indicated in gray; Ig, immunoglobulin (Ig)-like fold; TM, transmembrane segment; pre-Ig, the conserved segment between TM and Ig1. See Supplementary Figs. S1, S2 for details. b Limited chymotryptic digestion of full-length (FL) recombinant _Ct_pom152. c Negative-stain EM of chymotryptic fragment showed beaded and flexible strings of ~40 nm in length (37 ± 4 nm, number of particles: N = 30); two of the beaded strings (two dashed boxes) are shown enlarged (orange, bottom inserts); up to ten beads are discernible. d SDS-PAGE and Coomassie Blue staining of purified recombinant _Ct_pom152186-1270. e Like c, negative-stain EM of recombinant _Ct_pom152186-1270 also showed beaded string structures (measures 40 ± 5 nm, N = 30). f SEC-MALS indicates that _Ct_pom152186-1270 is a monomer. g–j Cryo-electron micrograph of _Ct_pom152FL. g Single particle showed flexible beaded strings (measures 44 ± 3 nm, N = 30), as seen in negative-stain EM (c, e) and four selected particles (orange with dashed box) are enlarged (h). i Two selected particles with high contrast showing ten beads (indicated by arrow heads) and the large structural variations. j _Ct_pom152FL polymerizes into long continuous strings with no punctuation marks. Particles represent the polymer formed by seven (bottom left), two (top), and five copies (right). k We speculate that (1) eight trans region of pom152 head-to-tail connect into a flexible ring and (2) two anti-parallel, stacked rings form above (cyan, omitted regions represented by dashed line), below (orange, omitted regions represented by dashed line), and mid-plane (black dashed line)
Similar articles
- Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J. Upla P, et al. Structure. 2017 Mar 7;25(3):434-445. doi: 10.1016/j.str.2017.01.006. Epub 2017 Feb 2. Structure. 2017. PMID: 28162953 Free PMC article. - The control of beads diameter of bead-on-string electrospun nanofibers and the corresponding release behaviors of embedded drugs.
Li T, Ding X, Tian L, Hu J, Yang X, Ramakrishna S. Li T, et al. Mater Sci Eng C Mater Biol Appl. 2017 May 1;74:471-477. doi: 10.1016/j.msec.2016.12.050. Epub 2016 Dec 13. Mater Sci Eng C Mater Biol Appl. 2017. PMID: 28254320 - Beads, beaded-fibres and fibres: Tailoring the morphology of poly(caprolactone) using pressurised gyration.
Hong X, Edirisinghe M, Mahalingam S. Hong X, et al. Mater Sci Eng C Mater Biol Appl. 2016 Dec 1;69:1373-82. doi: 10.1016/j.msec.2016.07.071. Epub 2016 Jul 28. Mater Sci Eng C Mater Biol Appl. 2016. PMID: 27612839 - Invasive pulmonary mycosis due to Chaetomium globosum with false-positive galactomannan test: a case report and literature review.
Capoor MR, Agarwal P, Goel M, Jain S, Shivaprakash MR, Honnavar P, Gupta S, Chakrabarti A. Capoor MR, et al. Mycoses. 2016 Mar;59(3):186-93. doi: 10.1111/myc.12446. Epub 2015 Dec 22. Mycoses. 2016. PMID: 26691935 Review. - [Aspergillus fumigatus and Chaetomium homopilatum in a leukemic patient. Pathogenic significance of Chaetomium species].
Schulze H, Aptroot A, Grote-Metke A, Balleisen L. Schulze H, et al. Mycoses. 1997;40 Suppl 1:104-9. doi: 10.1111/j.1439-0507.1997.tb00551.x. Mycoses. 1997. PMID: 9417506 Review. German.
Cited by
- Implications of a multiscale structure of the yeast nuclear pore complex.
Akey CW, Echeverria I, Ouch C, Nudelman I, Shi Y, Wang J, Chait BT, Sali A, Fernandez-Martinez J, Rout MP. Akey CW, et al. Mol Cell. 2023 Sep 21;83(18):3283-3302.e5. doi: 10.1016/j.molcel.2023.08.025. Mol Cell. 2023. PMID: 37738963 Free PMC article. - Characterization of nuclear pore complex targeting domains in Pom152 in Saccharomyces cerevisiae.
Brown JT, Haraczy AJ, Wilhelm CM, Belanger KD. Brown JT, et al. Biol Open. 2021 Oct 15;10(10):bio057661. doi: 10.1242/bio.057661. Epub 2021 Oct 20. Biol Open. 2021. PMID: 34557894 Free PMC article. - Lipid saturation controls nuclear envelope function.
Romanauska A, Köhler A. Romanauska A, et al. Nat Cell Biol. 2023 Sep;25(9):1290-1302. doi: 10.1038/s41556-023-01207-8. Epub 2023 Aug 17. Nat Cell Biol. 2023. PMID: 37591950 Free PMC article. - The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth.
Dultz E, Wojtynek M, Medalia O, Onischenko E. Dultz E, et al. Cells. 2022 Apr 25;11(9):1456. doi: 10.3390/cells11091456. Cells. 2022. PMID: 35563762 Free PMC article. Review. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources