Base editing: precision chemistry on the genome and transcriptome of living cells - PubMed (original) (raw)
Review
Base editing: precision chemistry on the genome and transcriptome of living cells
Holly A Rees et al. Nat Rev Genet. 2018 Dec.
Erratum in
- Publisher Correction: Base editing: precision chemistry on the genome and transcriptome of living cells.
Rees HA, Liu DR. Rees HA, et al. Nat Rev Genet. 2018 Dec;19(12):801. doi: 10.1038/s41576-018-0068-0. Nat Rev Genet. 2018. PMID: 30341440
Abstract
RNA-guided programmable nucleases from CRISPR systems generate precise breaks in DNA or RNA at specified positions. In cells, this activity can lead to changes in DNA sequence or RNA transcript abundance. Base editing is a newer genome-editing approach that uses components from CRISPR systems together with other enzymes to directly install point mutations into cellular DNA or RNA without making double-stranded DNA breaks. DNA base editors comprise a catalytically disabled nuclease fused to a nucleobase deaminase enzyme and, in some cases, a DNA glycosylase inhibitor. RNA base editors achieve analogous changes using components that target RNA. Base editors directly convert one base or base pair into another, enabling the efficient installation of point mutations in non-dividing cells without generating excess undesired editing by-products. In this Review, we summarize base-editing strategies to generate specific and precise point mutations in genomic DNA and RNA, highlight recent developments that expand the scope, specificity, precision and in vivo delivery of base editors and discuss limitations and future directions of base editing for research and therapeutic applications.
Figures
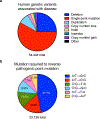
Figure 1. Distribution of human pathogenic genetic variants, including point mutations.
a) Classification of human pathogenic genetic variants in the ClinVar database (accessed May 29, 2018),. As noted in the text, sampling bias due to the extensive use of short-read sequencing to analyze genomic diversity is possible. b) Distribution of base pair changes needed to reverse the pathogenic point mutations represented in the red wedge in (a),. The percentage represented by each base pair change is noted on the pie chart; transition mutations are labeled in white and transversion mutations are labeled in black.
Figure 2. Cytosine base editing.
(a) Cytosine deamination generates uracil, which base pairs as thymidine. R = 2′-deoxyribose in DNA, or ribose in RNA. (b) Cytosine base editing strategy by BE1, BE2, BE3, or BE4. R-loop formation exposes a region of single-stranded DNA to the cytidine deaminase domain. Target cytosines in this region are deaminated to uracil,. (c) Cellular response to cytosine base editing. Uracil DNA glycosylase-mediated excision of the uracil generated in genomic DNA is inhibited by BE2, BE3, and BE4. BE3 and BE4 are designed to nick the non-edited strand (containing the G of the original C•G target base pair), stimulating cellular DNA repair of that strand to replace the G with an A, completing the conversion of the original C•G base pair to a U•A or, following DNA replication or repair to a T•A base pair,.
Figure 3. Adenine base editing in DNA and RNA.
(a) Adenosine deamination generates inosine, which has the same base pairing preferences as a guanosine in the active site of a polymerase. R = 2′-deoxyribose in DNA, or ribose in RNA. (b) ABE-mediated DNA base editing to convert an A•T base pair to a G•C base pair. Current ABEs contain one wild-type TadA structural monomer and one evolved TadA catalytic monomer. R-loop formation exposes a small region of ssDNA, within which A is deaminated to I by the heterodimeric wild-type TadA–evolved TadA heterodimer.(c-e) Antisense-RNA mediated RNA editing. ADAR variants are localized to the RNA transcript of interest through an antisense RNA with variable lengths of homology to the target transcript. The target A is specified with an A•C mismatch in the mRNA:antisense-RNA duplex. The antisense RNA is in orange, and the target mRNA is in black. (c) Antisense-directed RNA editing by covalent linkage of an ADAR deaminase domain (ADARDD)–SNAP tag fusion to a benzylguanine (BzG)-modified antisense RNA,,. (d) ADAR-BoxB mediated RNA editing,. The ADAR2DD is fused to λN protein; λN binds to one of the two BoxB hairpins integrated into the antisense RNA, localizing ADAR-mediated deamination activity to the target adenine. (e) Full-length ADAR2-mediated RNA editing. The antisense RNA comprises a 5’ R/G-binding motif hairpin, the native binding sequence for full length ADAR2, followed by a 19-nt antisense region complementary to the target RNA with the target adenine centrally located and specified by an A•C mismatch. Overexpression of full-length ADAR2 results in localization of the deaminase to the target base within the target transcript through the native ADAR2 double-stranded RNA binding domains. (f) RNA editing with REPAIR. A dCas13b:guide RNA complex is guided to a the target RNA by a 50-nt spacer. The target A is specified by an A•C mismatch centrally located within the 50-nt spacer..
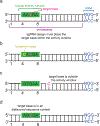
Figure 4. Overcoming targeting challenges associated with base editing.
An example of a cytosine base editing site has been shown; these principles also apply to other classes of base editors (a) An ideal base editing target locus. The target base is located within the base editor activity window relative to the PAM site, there is only one target base in the activity window, and the target base is found in a motif (AC in this example) that is efficiently deaminated by most cytosine base editors,. (b) Example of a target site with a bystander base. If the bystander edit (deamination of the cytosine shown in yellow) is undesired, a narrowed-window or context-specific base editor may be used to preferentially edit the target base over the bystander base (c) The target base is located outside of the activity window. Base editing on this target may be possible with base editors that recognize different PAMs (see Table 1). (d) The target base is located within a sequence context that may not an efficient substrate for a particular deaminase,. Editing of the target may be improved by using an editor with a different deaminase,, or an editor more tolerant of methylated DNA.
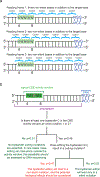
“Figure 5” (Figure to accompany Box 1)
Illustration of the probability calculations for non-silent bystander editing in a protein-coding gene, as described in Box 1. (a) Use of base editing to generate a non-silent mutation in the protein-coding region of a gene. There are three possible orientations of the base editor activity window relative to the reading frame of the protein. Base editors currently generate transition mutations, and the vast majority (97%) of transitions at codon position 3 are silent. Thus, the target base in a coding gene is predominantly located at coding position 1 or 2 (N1 or N2), because the consequence of generating a transition at N3 is almost always to generate a silent mutation. In a 5-nt activity window, typical of a CBE, an average of 2.33 nucleotides at positions N1 or N2 are present in the window, in addition to the target nucleotide. For a 4-nt window typical of an ABE there are on average 1.75 nucleotides at codon positions 1 or 2 in addition to the target nucleotide (see Box 1). These additional editable nucleotides at N1 or N2, depending on their identity and sequence context, may contribute to bystander editing (see Box 1). (b) Flow chart to calculate the probability of finding a bystander mutation within a 5-nt window, as described in Box 1. The probability of the indicated event (p) assumes a genome with randomly distributed bases. For a description of how each probability was calculated, see Box 1. The same calculation is described for a 4-nt window typical of ABEs in Box 1.
Similar articles
- Editing the Genome Without Double-Stranded DNA Breaks.
Komor AC, Badran AH, Liu DR. Komor AC, et al. ACS Chem Biol. 2018 Feb 16;13(2):383-388. doi: 10.1021/acschembio.7b00710. Epub 2017 Oct 9. ACS Chem Biol. 2018. PMID: 28957631 Free PMC article. Review. - Off-Target Editing by CRISPR-Guided DNA Base Editors.
Park S, Beal PA. Park S, et al. Biochemistry. 2019 Sep 10;58(36):3727-3734. doi: 10.1021/acs.biochem.9b00573. Epub 2019 Aug 26. Biochemistry. 2019. PMID: 31433621 Free PMC article. Review. - Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors.
Anzalone AV, Koblan LW, Liu DR. Anzalone AV, et al. Nat Biotechnol. 2020 Jul;38(7):824-844. doi: 10.1038/s41587-020-0561-9. Epub 2020 Jun 22. Nat Biotechnol. 2020. PMID: 32572269 Review. - Current Status and Challenges of DNA Base Editing Tools.
Jeong YK, Song B, Bae S. Jeong YK, et al. Mol Ther. 2020 Sep 2;28(9):1938-1952. doi: 10.1016/j.ymthe.2020.07.021. Epub 2020 Jul 23. Mol Ther. 2020. PMID: 32763143 Free PMC article. Review. - Prime editing for precise and highly versatile genome manipulation.
Chen PJ, Liu DR. Chen PJ, et al. Nat Rev Genet. 2023 Mar;24(3):161-177. doi: 10.1038/s41576-022-00541-1. Epub 2022 Nov 7. Nat Rev Genet. 2023. PMID: 36344749 Free PMC article. Review.
Cited by
- Advances in genomic tools for plant breeding: harnessing DNA molecular markers, genomic selection, and genome editing.
Kumar R, Das SP, Choudhury BU, Kumar A, Prakash NR, Verma R, Chakraborti M, Devi AG, Bhattacharjee B, Das R, Das B, Devi HL, Das B, Rawat S, Mishra VK. Kumar R, et al. Biol Res. 2024 Nov 7;57(1):80. doi: 10.1186/s40659-024-00562-6. Biol Res. 2024. PMID: 39506826 Review. - The design and engineering of synthetic genomes.
James JS, Dai J, Chew WL, Cai Y. James JS, et al. Nat Rev Genet. 2024 Nov 6. doi: 10.1038/s41576-024-00786-y. Online ahead of print. Nat Rev Genet. 2024. PMID: 39506144 Review. - Applications of CRISPR/Cas as a Toolbox for Hepatitis B Virus Detection and Therapeutics.
Kumar A, Combe E, Mougené L, Zoulim F, Testoni B. Kumar A, et al. Viruses. 2024 Oct 2;16(10):1565. doi: 10.3390/v16101565. Viruses. 2024. PMID: 39459899 Free PMC article. Review. - Leveraging CRISPR gene editing technology to optimize the efficacy, safety and accessibility of CAR T-cell therapy.
Lei T, Wang Y, Zhang Y, Yang Y, Cao J, Huang J, Chen J, Chen H, Zhang J, Wang L, Xu X, Gale RP, Wang L. Lei T, et al. Leukemia. 2024 Oct 25. doi: 10.1038/s41375-024-02444-y. Online ahead of print. Leukemia. 2024. PMID: 39455854 Review.
References
- Jansen R, Embden JD, Gaastra W & Schouls LM Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43, 1565–1575 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RM1 HG009490/HG/NHGRI NIH HHS/United States
- R35 GM118062/GM/NIGMS NIH HHS/United States
- R01 GM065400/GM/NIGMS NIH HHS/United States
- R01 GM065865/GM/NIGMS NIH HHS/United States
- R01 EB022376/EB/NIBIB NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources