Victors: a web-based knowledge base of virulence factors in human and animal pathogens - PubMed (original) (raw)
. 2019 Jan 8;47(D1):D693-D700.
doi: 10.1093/nar/gky999.
Li Li 2, Edison Ong 1, Shunzhou Deng 1 3, Guanghua Fu 1 4, Yu Lin 1, Brian Yang 1, Shelley Zhang 1, Zhenzong Fa 5, Bin Zhao 1, Zuoshuang Xiang 1, Yongqing Li 6, Xing-Ming Zhao 7, Michal A Olszewski 5, Luonan Chen 2 8 9, Yongqun He 1
Affiliations
- PMID: 30365026
- PMCID: PMC6324020
- DOI: 10.1093/nar/gky999
Victors: a web-based knowledge base of virulence factors in human and animal pathogens
Samantha Sayers et al. Nucleic Acids Res. 2019.
Abstract
Virulence factors (VFs) are molecules that allow microbial pathogens to overcome host defense mechanisms and cause disease in a host. It is critical to study VFs for better understanding microbial pathogenesis and host defense mechanisms. Victors (http://www.phidias.us/victors) is a novel, manually curated, web-based integrative knowledge base and analysis resource for VFs of pathogens that cause infectious diseases in human and animals. Currently, Victors contains 5296 VFs obtained via manual annotation from peer-reviewed publications, with 4648, 179, 105 and 364 VFs originating from 51 bacterial, 54 viral, 13 parasitic and 8 fungal species, respectively. Our data analysis identified many VF-specific patterns. Within the global VF pool, cytoplasmic proteins were more common, while adhesins were less common compared to findings on protective vaccine antigens. Many VFs showed homology with host proteins and the human proteins interacting with VFs represented the hubs of human-pathogen interactions. All Victors data are queriable with a user-friendly web interface. The VFs can also be searched by a customized BLAST sequence similarity searching program. These VFs and their interactions with the host are represented in a machine-readable Ontology of Host-Pathogen Interactions. Victors supports the 'One Health' research as a vital source of VFs in human and animal pathogens.
Figures
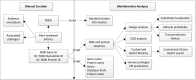
Figure 1.
The workflow of the Victors virulence factor annotation and analysis. .
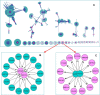
Figure 2.
Landscapes of predicted bacteria pathogen–human PPIs. (A) All 15 334 PPIs. (B) This figure shows that two pathogen proteins (GPT pyrophosphokinase rsh) from Brucella suis (Q8CY42) and Brucella melitensis (Q8YG65) interact with a group of human proteins (blue dots) on tRNA level, such as t-RNA editing proteins, tRNA ligase proteins. The GO enrichment analysis reveals that this group of proteins is enriched in tRNA aminoacylation (A2RTX5|Q9BW92|Q5JTZ9|P49588|P26639), corrected _P_-value 9.5487E-12. (C) This figure shows that two human proteins lengsin (Q5TDP6) and Glutamine synthetase (P15104) interact with a group of pathogen proteins from different species, such as Streptococcus pneumonia and Brucella suis, Brucella melitensis. Interestingly, all pathogen proteins are Glutamine synthetase. Nodes in red color are pathogen proteins, and nodes in blue are human proteins.
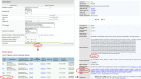
Figure 3.
Example of Victors data query and BLAST sequence similarity analysis. In this example, a user selected ‘_Mycobacterium tuberculosis_’ in the ‘Pathogen or Disease’ menu bar and typed ‘transcriptional regulatory protein’. After clicking the ‘Search’ button, a list of 19 genes was shown up in the results page. A click on the Victors ID ‘110’ (the forth on the list) led to another page providing details about the VF (e.g. the gene mutation phenotype information and predicted interacting host proteins). A BLAST search was initiated by clicking ‘BLAST’.
Similar articles
- VFDB 2019: a comparative pathogenomic platform with an interactive web interface.
Liu B, Zheng D, Jin Q, Chen L, Yang J. Liu B, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D687-D692. doi: 10.1093/nar/gky1080. Nucleic Acids Res. 2019. PMID: 30395255 Free PMC article. - Common and pathogen-specific virulence factors are different in function and structure.
Niu C, Yu D, Wang Y, Ren H, Jin Y, Zhou W, Li B, Cheng Y, Yue J, Gao Z, Liang L. Niu C, et al. Virulence. 2013 Aug 15;4(6):473-82. doi: 10.4161/viru.25730. Epub 2013 Jul 15. Virulence. 2013. PMID: 23863604 Free PMC article. - VFDB 2022: a general classification scheme for bacterial virulence factors.
Liu B, Zheng D, Zhou S, Chen L, Yang J. Liu B, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D912-D917. doi: 10.1093/nar/gkab1107. Nucleic Acids Res. 2022. PMID: 34850947 Free PMC article. - Opportunistic yeast pathogen Candida spp.: Secreted and membrane-bound virulence factors.
Lim SJ, Mohamad Ali MS, Sabri S, Muhd Noor ND, Salleh AB, Oslan SN. Lim SJ, et al. Med Mycol. 2021 Dec 3;59(12):1127-1144. doi: 10.1093/mmy/myab053. Med Mycol. 2021. PMID: 34506621 Review. - Unravelling bacterial virulence factors in yeast: From identification to the elucidation of their mechanisms of action.
Wong Z, Ong EBB. Wong Z, et al. Arch Microbiol. 2024 Jun 15;206(7):303. doi: 10.1007/s00203-024-04023-2. Arch Microbiol. 2024. PMID: 38878203 Review.
Cited by
- Genomic population structure of Helicobacter pylori Shanghai isolates and identification of genomic features uniquely linked with pathogenicity.
Yang F, Zhang J, Wang S, Sun Z, Zhou J, Li F, Liu Y, Ding L, Liu Y, Chi W, Liu T, He Y, Xiang P, Bao Z, Olszewski MA, Zhao H, Zhang Y. Yang F, et al. Virulence. 2021 Dec;12(1):1258-1270. doi: 10.1080/21505594.2021.1920762. Virulence. 2021. PMID: 33904371 Free PMC article. - Culturable Bacterial Endophytes of Wild White Poplar (Populus alba L.) Roots: A First Insight into Their Plant Growth-Stimulating and Bioaugmentation Potential.
Gladysh NS, Bogdanova AS, Kovalev MA, Krasnov GS, Volodin VV, Shuvalova AI, Ivanov NV, Popchenko MI, Samoilova AD, Polyakova AN, Dmitriev AA, Melnikova NV, Karpov DS, Bolsheva NL, Fedorova MS, Kudryavtseva AV. Gladysh NS, et al. Biology (Basel). 2023 Dec 12;12(12):1519. doi: 10.3390/biology12121519. Biology (Basel). 2023. PMID: 38132345 Free PMC article. - Association of pathogenic determinants of Fusobacterium necrophorum with bacteremia, and Lemierre's syndrome.
Carrara A, Bertelli C, Gardiol C, Marquis B, Andrey DO, Schrenzel J, Pillonel T, Greub G. Carrara A, et al. Sci Rep. 2024 Aug 27;14(1):19804. doi: 10.1038/s41598-024-70608-y. Sci Rep. 2024. PMID: 39191804 Free PMC article. - Scalable and versatile container-based pipelines for de novo genome assembly and bacterial annotation.
de Almeida FM, de Campos TA, Pappas GJ Jr. de Almeida FM, et al. F1000Res. 2023 Sep 25;12:1205. doi: 10.12688/f1000research.139488.1. eCollection 2023. F1000Res. 2023. PMID: 37970066 Free PMC article. Review. - Compounding Achromobacter Phages for Therapeutic Applications.
Cobián Güemes AG, Le T, Rojas MI, Jacobson NE, Villela H, McNair K, Hung SH, Han L, Boling L, Octavio JC, Dominguez L, Cantú VA, Archdeacon S, Vega AA, An MA, Hajama H, Burkeen G, Edwards RA, Conrad DJ, Rohwer F, Segall AM. Cobián Güemes AG, et al. Viruses. 2023 Jul 30;15(8):1665. doi: 10.3390/v15081665. Viruses. 2023. PMID: 37632008 Free PMC article.
References
- Atlas R.M. One Health: its origins and future. Curr. Top. Microbiol. Immunol. 2013; 365:1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials