Complete genome analysis of a SARS-like bat coronavirus identified in the Republic of Korea - PubMed (original) (raw)
Complete genome analysis of a SARS-like bat coronavirus identified in the Republic of Korea
Yongkwan Kim et al. Virus Genes. 2019 Aug.
Abstract
Bats have been widely known as natural reservoir hosts of zoonotic diseases, such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) caused by coronaviruses (CoVs). In the present study, we investigated the whole genomic sequence of a SARS-like bat CoV (16BO133) and found it to be 29,075 nt in length with a 40.9% G+C content. Phylogenetic analysis using amino acid sequences of the ORF 1ab and the spike gene showed that the bat coronavirus strain 16BO133 was grouped with the Beta-CoV lineage B and was closely related to the JTMC15 strain isolated from Rhinolophus ferrumequinum in China. However, 16BO133 was distinctly located in the phylogenetic topology of the human SARS CoV strain (Tor2). Interestingly, 16BO133 showed complete elimination of ORF8 regions induced by a frame shift of the stop codon in ORF7b. The lowest amino acid identity of 16BO133 was identified at the spike region among various ORFs. The spike region of 16BO133 showed 84.7% and 75.2% amino acid identity with Rf1 (SARS-like bat CoV) and Tor2 (human SARS CoV), respectively. In addition, the S gene of 16BO133 was found to contain the amino acid substitution of two critical residues (N479S and T487 V) associated with human infection. In conclusion, we firstly carried out whole genome characterization of the SARS-like bat coronavirus discovered in the Republic of Korea; however, it presumably has no human infectivity. However, continuous surveillance and genomic characterization of coronaviruses from bats are necessary due to potential risks of human infection induced by genetic mutation.
Keywords: Bat; Frame shift; SARS-like coronavirus; Whole genome; Zoonotic disease.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
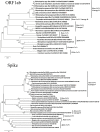
Fig. 1
Phylogenetic analysis using whole genome sequences of ORF 1ab with reference strains. The phylogenetic trees were drawn using the neighboring joining method using the maximum composite likelihood model with MEGA 7 software. The bootstrap values were calculated with 1000 replicates. Phylogenetic analysis using whole genome sequences of the spike region with reference strains. The phylogenetic trees were drawn using the neighboring joining method using the maximum composite likelihood model with MEGA 7 software. The bootstrap values were calculated with 1000 replicates
Similar articles
- Comparative genetic analyses of Korean bat coronaviruses with SARS-CoV and the newly emerged SARS-CoV-2.
Na EJ, Lee SY, Kim HJ, Oem JK. Na EJ, et al. J Vet Sci. 2021 Jan;22(1):e12. doi: 10.4142/jvs.2021.22.e12. J Vet Sci. 2021. PMID: 33522164 Free PMC article. - Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.
Lau SK, Feng Y, Chen H, Luk HK, Yang WH, Li KS, Zhang YZ, Huang Y, Song ZZ, Chow WN, Fan RY, Ahmed SS, Yeung HC, Lam CS, Cai JP, Wong SS, Chan JF, Yuen KY, Zhang HL, Woo PC. Lau SK, et al. J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269185 Free PMC article. - Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.
Li L, Zhang L, Zhou J, He X, Yu Y, Liu P, Huang W, Xiang Z, Chen J. Li L, et al. mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article. - Molecular epidemiology, evolution and phylogeny of SARS coronavirus.
Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Luk HKH, et al. Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review. - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
- A comprehensive dataset of animal-associated sarbecoviruses.
Liu B, Zhao P, Xu P, Han Y, Wang Y, Chen L, Wu Z, Yang J. Liu B, et al. Sci Data. 2023 Oct 7;10(1):681. doi: 10.1038/s41597-023-02558-5. Sci Data. 2023. PMID: 37805633 Free PMC article. - Genotype and Phenotype Characterization of Rhinolophus sp. Sarbecoviruses from Vietnam: Implications for Coronavirus Emergence.
Temmam S, Tu TC, Regnault B, Bonomi M, Chrétien D, Vendramini L, Duong TN, Phong TV, Yen NT, Anh HN, Son TH, Anh PT, Amara F, Bigot T, Munier S, Thong VD, van der Werf S, Nam VS, Eloit M. Temmam S, et al. Viruses. 2023 Sep 8;15(9):1897. doi: 10.3390/v15091897. Viruses. 2023. PMID: 37766303 Free PMC article. - High activity of an affinity-matured ACE2 decoy against Omicron SARS-CoV-2 and pre-emergent coronaviruses.
Sims JJ, Lian S, Meggersee RL, Kasimsetty A, Wilson JM. Sims JJ, et al. PLoS One. 2022 Aug 25;17(8):e0271359. doi: 10.1371/journal.pone.0271359. eCollection 2022. PLoS One. 2022. PMID: 36006993 Free PMC article. - Genomic Comparisons of Alphacoronaviruses and Betacoronaviruses from Korean Bats.
Lo VT, Yoon SW, Choi YG, Jeong DG, Kim HK. Lo VT, et al. Viruses. 2022 Jun 25;14(7):1389. doi: 10.3390/v14071389. Viruses. 2022. PMID: 35891370 Free PMC article. - SARS-CoV-2 in animals: potential for unknown reservoir hosts and public health implications.
Sharun K, Dhama K, Pawde AM, Gortázar C, Tiwari R, Bonilla-Aldana DK, Rodriguez-Morales AJ, de la Fuente J, Michalak I, Attia YA. Sharun K, et al. Vet Q. 2021 Dec;41(1):181-201. doi: 10.1080/01652176.2021.1921311. Vet Q. 2021. PMID: 33892621 Free PMC article. Review.
References
- Kim HK, Yoon SW, Kim DJ, Koo BS, Noh JY, Kim JH, Choi YG, Na W, Chang KT, Song D, Jeong DG. Detection of severe acute respiratory syndrome-like, Middle East respiratory syndrome-like bat coronaviruses and group H rotavirus in faeces of Korean bats. Transbound Emerg Dis. 2016;63:365–372. doi: 10.1111/tbed.12515. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous