MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis - PubMed (original) (raw)
MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis
Qing Ji et al. Cell Death Dis. 2019.
Abstract
Ectopic expression of lncRNA-MALAT1 has been discovered in recurrent colorectal cancer (CRC) and metastatic sites in postsurgical patients, however, its biological mechanism remained unelucidated. Our study first revealed the novel roles of MALAT1 in promoting CRC metastasis through two mechanisms: first, MALAT1 binds miR-15 family members, to "de-inhibit" their effect on LRP6 expression, enhances β-catenin signaling, leading to elevated transcriptional levels of downstream target genes RUNX2. Second, MALAT1 binds SFPQ, and dissociates SFPQ/PTBP2 dimer to release free PTBP2, which elevates translational levels of RUNX2, through interacting with IRES domain in the 5'UTR of the corresponding RUNX2 mRNAs. Moreover, increased RUNX2 expression levels were detected in recurrent CRC tumors, which were closely associated with TMN stages, metastasis, as well as CRC patients' survival. Our study demonstrated that MALAT1 and RUNX2 may serve as two biomarkers for predicting the recurrence and metastasis of CRC patients.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
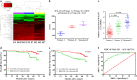
Fig. 1. MALAT1 expression in postsurgical, recurrent primary, and metastatic sites, compared with primary sites of non-recurrent CRC patients.
a Cluster analysis of differentially expressed lncRNAs in three CRC primary tumor tissues of non-recurrent patients (P4, P8, and P10), and three paired tissues (primary P2, P3, and P7, and metastatic M2, M3, and M7) of recurrent CRC patients. Red color represents high expression and green color represents low expression. The color brightness of each unit is associated with differences in multiples (log 2(AR/N). Not all the miRNAs in the figure were labeled. b Expression of MALAT1 in three CRC primary tissues and three paired tissues, including CRC primary tissues and metastatic tissues (nine samples for RNA sequencing). FPKM in the Y axis represents fragments per kilobase of exon per million fragments mapped. c Expression levels of MALAT1 in 124 CRC tissues and matched metastatic sites were analyzed by qRT-PCR. The significant differences between primary tumor I (without paired metastatic tissues) and primary tumor II (with paired metastatic tissues, Metastasis II) were analyzed using the Wilcoxon signed-rank test. d–e Kaplan–Meier analyses of the correlations between MALAT1 expression levels and overall survival (OS) and disease-free survival (DFS) of 124 CRC patients, and the median expression level was used as the cutoff. f A ROC curve of CRC patients based on MALAT1 expression in primary tumor I and primary tumor II. *P < 0.05; **P < 0.01 (t test)
Fig. 2. CRISPR/Cas9-mediated MALAT1 knockout elevates the levels of miR-15s.
a Two CRISPR nuclease sgRNA designs for lncRNA-MALAT1. b Functional validation in HEK293T cells by SURVEYOR Nuclease S Assay. c Validation of CRISPR knockout in single-cell clones by PCR and DNA sequencing. d Northern blot analysis of MALAT1 expression. Twenty micrograms of total RNA from MALAT1 KO LoVo cells (M4) and wild-type LoVo cells were run in agarose/formaldehyde gels. After transferring to nylon membranes, MALAT1 and GAPDH mRNA were detected using antisense oligomer probes (MALAT1 KO Probe 1 targeting the deleted core region, MALAT1 KO Probe 2 targeting the deleted non-core region, and control GAPDH probe) that were end-labeled with 32P. e Cluster analysis of differentially expressed miRNAs in LoVo cells and LoVo/MALAT1−/− cells following by RNA sequencing. Red color represents high expression and green color represents low expression. The color brightness of each unit is associated with differences in multiples (log 2(AR/N). Not all the miRNAs in the figure were labeled. f Validation of the miR-15s levels in LoVo/MALAT1–/–cells by real-time PCR assay. *P < 0.05; **P < 0.01 (t test)
Fig. 3. MALAT1 functions as a ceRNA regulating miR-15 family.
a Interaction network diagram between MALAT1 and differentially expressed miRNAs characterized by RNA sequencing using star-Base v2.0, microRNA.org, and MirTarget2. b–c MS2-RIP assay followed by real-time PCR to detect miRNAs endogenously associated with MALAT1. d LoVo cell lysates were incubated with biotin-labeled MALAT1. After pulldown, the whole miRNAs were extracted and measured by real-time PCR. e RIP analysis of the interaction between Ago2 and MALAT1 using IgG and antibodies recognizing AGO2. MALAT1 and housekeeping GAPDH mRNA abundance was quantified using real-time PCR and represented as enrichment in RBP RIP compared with IgG RIP. f Anti-AGO2 RIP was performed in LoVo cells transiently overexpressing miR-15s, followed by qRT-PCR to detect MALAT1 or MALAT-mut (miR-15s) associated with AGO2. g Forty-eight hours after silencing AGO2 in LoVo cells, MS2-RIP assay followed by real-time PCR was used to detect miRNAs endogenously associated with MALAT1. h Forty-eight hours after silencing AGO2 in LoVo cells, cells were lysed and the cell lysates were incubated with biotin-labeled MALAT1. After pulldown, the whole miRNAs were extracted and measured by real-time PCR. i Levels of miR-15s in MALAT1 knockout LoVo cells. *P < 0.05; **P < 0.01 (t test)
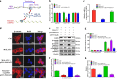
Fig. 4. MALAT1 promotes the RUNX2 transcription through LRP6-mediated β-catenin signaling pathway.
a Schematic diagram of the interlink between MALAT1 and miR-15s or between miR-15s and LRP6. b Luciferase reporter activities of wild-type mutant LRP6 3′UTR reporter, and empty constructs in LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. c Real-time PCR assay of LRP6 mRNA levels in LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. d Immunofluorescence detection of β-catenin LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. e–f Western blot and quantitative assay of β-catenin (nuclear) in LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. g LEF/TCF promoter activity assay in LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. h Real-time PCR assay of RUNX2 mRNA levels in LoVo/MALAT1−/− cells and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. XAV939, the inhibitor for β-catenin signaling pathway, was used to block the activation of β-catenin signaling pathway and observe whether there was any effect of MALAT1 through the β-catenin signaling pathway. *P < 0.05; **P < 0.01 (t test)
Fig. 5. PTBP2 regulates the translation of RUNX2 in CRC cells.
a Insertion of the RUNX2, VEGFa, and COX-2 5′UTR sequences (410 bp, 1038 bp, and 134 bp, respectively) into the empty dicistronic vector pR-F. b Renilla (RL) and firefly (FL) luciferase activity analyses following transfection of LoVo cells with different concentrations (0 ng, 12.5 ng, 25 ng, and 50 ng) of PTBP2-expressing plasmid. c Biotinylated RUNX2, VEGFa, and COX-2 5′UTR RNAs bound to streptavidin beads were incubated with cytoplasmic extracts from LoVo cells. After extensive washing, RNA-bound proteins were analyzed by western blot using an anti-PTBP2 antibody. d Schematic representation of the RUNX2 and VEGFa fragments. The polypyrimidine tract (PPT) is indicated as a black ellipse. e–f Biotinylated RUNX2 and VEGFa 5′UTR RNAs or fragment RNAs bound to streptavidin beads were incubated with cytoplasmic extracts from LoVo cells. After extensive washing, RNA-bound proteins were analyzed by western blot using an anti-PTBP2 antibody. g–j qPCR and western blot analysis for RUNX2, VEGFa, and COX-2 in LoVo and SW620 CRC cells transfected with PTBP2-expressing plasmid
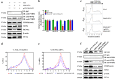
Fig. 6. MALAT1 associates with polysomes and promotes RUNX2 translation.
a Western blot and quantitative assay of free PTBP-2 protein in LoVo, LoVo/MALAT1−/−, and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. b Renilla (RL) and firefly (FL) luciferase activity analyses following transfection of pR-F, pRrunx2F, pRVEGFaF, or pRcox-2F in LoVo, LoVo/MALAT1−/−, and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells. c Polysomes in cytoplasmic extracts from LoVo, LoVo/MALAT1−/−, and pLV4-MALAT1-transfected LoVo/MALAT1−/− cells were fractionated through sucrose gradients (arrow: direction of sedimentation; –, no ribosomal components). d–e Relative distributions of MALAT1 lncRNA and RUNX2 mRNAs were analyzed by real-time PCR of RNA in gradient fractions, and represented as percent of total RNA in the gradient. f Western blot assay for detecting the effect of PTBP2 depletion on the MALAT1’s capability that regulated the level of RUNX2
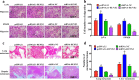
Fig. 7. RUNX2 promotes the metastasis of CRC cells in vitro and in vivo.
a Migration assays of LoVo or SW620 cells transfected with pcDNA3.1, pcDNA3.1-RUNX2, shRNA-NC, and shRNA–RUNX2, respectively. b Numbers of migrated cells were shown as mean ± SD; n = 3. c Representative results of H&E staining of the metastatic nodules in the lung and liver from six mice subjected to the indicated treatments. d Quantification of lung and liver metastasis from six mice subjected to the indicated treatments. *P < 0.05; **P < 0.01 (t test)
Fig. 8. RUNX2 overexpression correlates with CRC progression.
a Expression levels of RUNX2 in CRC tissues were analyzed by real-time PCR. The significant differences between primary tumor I (without paired metastatic tissues) and primary tumor II (with paired metastatic tissues) were analyzed using the Wilcoxon signed-rank test. **P < 0.01 (t test). b–c Kaplan–Meier analyses of the correlations between RUNX2 expression levels and OS or DFS of 124 CRC patients; the median expression levels were used as the cutoff. d Immunohistochemical analysis of RUNX2 proteins on consecutive tissue microarray slides of CRC tissues. Representative case 1 and case 2 were presented (scale bars, 200 and 50 mm, respectively). e A schematic model of MALAT1 regulating transcriptional and translational levels of RUNX2 in CRC metastasis
Similar articles
- Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex.
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Ji Q, et al. Br J Cancer. 2014 Aug 12;111(4):736-48. doi: 10.1038/bjc.2014.383. Epub 2014 Jul 15. Br J Cancer. 2014. PMID: 25025966 Free PMC article. - Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway.
Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y, Li S, Qi Y, Huang Y, Jia L. Xu J, et al. J Exp Clin Cancer Res. 2020 Mar 24;39(1):54. doi: 10.1186/s13046-020-01562-6. J Exp Clin Cancer Res. 2020. PMID: 32209115 Free PMC article. - PTBP3 contributes to colorectal cancer growth and metastasis via translational activation of HIF-1α.
Hou P, Chen F, Yong H, Lin T, Li J, Pan Y, Jiang T, Li M, Chen Y, Song J, Zheng J, Bai J. Hou P, et al. J Exp Clin Cancer Res. 2019 Jul 10;38(1):301. doi: 10.1186/s13046-019-1312-y. J Exp Clin Cancer Res. 2019. PMID: 31291975 Free PMC article. - MALAT1: A Promising Therapeutic Target for the Treatment of Metastatic Colorectal Cancer.
Uthman YA, Ibrahim KG, Abubakar B, Bello MB, Malami I, Imam MU, Qusty N, Cruz-Martins N, Batiha GE, Abubakar MB. Uthman YA, et al. Biochem Pharmacol. 2021 Aug;190:114657. doi: 10.1016/j.bcp.2021.114657. Epub 2021 Jun 16. Biochem Pharmacol. 2021. PMID: 34144008 Review. - The Biological Effect and Clinical Application of Long Noncoding RNAs in Colorectal Cancer.
Sun Z, Liu J, Chen C, Zhou Q, Yang S, Wang G, Song J, Li Z, Zhang Z, Xu J, Sun X, Chang Y, Yuan W. Sun Z, et al. Cell Physiol Biochem. 2018;46(2):431-441. doi: 10.1159/000488610. Epub 2018 Mar 26. Cell Physiol Biochem. 2018. PMID: 29614491 Review.
Cited by
- YYFZBJS ameliorates colorectal cancer progression in ApcMin/+ mice by remodeling gut microbiota and inhibiting regulatory T-cell generation.
Sui H, Zhang L, Gu K, Chai N, Ji Q, Zhou L, Wang Y, Ren J, Yang L, Zhang B, Hu J, Li Q. Sui H, et al. Cell Commun Signal. 2020 Jul 16;18(1):113. doi: 10.1186/s12964-020-00596-9. Cell Commun Signal. 2020. PMID: 32677955 Free PMC article. - The long non-coding RNA MALAT1 regulates intestine host-microbe interactions and polyposis.
Long T, Hernandez JE, Ma S, Steele S, Luo C, Li Y, Xie Q, Telese F, Zhou B, Huang WJM. Long T, et al. Front Cell Dev Biol. 2023 May 30;11:1168693. doi: 10.3389/fcell.2023.1168693. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37325561 Free PMC article. - Unveiling the Intricacies of Monoamine Oxidase-A (MAO-A) Inhibition in Colorectal Cancer: Computational Systems Biology, Expression Patterns, and the Anticancer Therapeutic Potential.
Bardaweel SK, Al-Salamat H, Hajjo R, Sabbah D, Almutairi S. Bardaweel SK, et al. ACS Omega. 2024 Aug 9;9(33):35703-35717. doi: 10.1021/acsomega.4c04100. eCollection 2024 Aug 20. ACS Omega. 2024. PMID: 39184489 Free PMC article. - Long non-coding RNA AK001058 regulates tumor growth and angiogenesis in colorectal cancer via methylation of ADAMTS12.
Zheng S, Lin F, Zhang M, Mu N, Ge X, Fu J. Zheng S, et al. Am J Transl Res. 2019 Sep 15;11(9):6117-6123. eCollection 2019. Am J Transl Res. 2019. PMID: 31632580 Free PMC article. - Blocking long noncoding RNA MALAT1 restrained the development of laryngeal and hypopharyngeal carcinoma.
Xu E, Liang X, Ji Z, Zhao S, Li L, Lang J. Xu E, et al. Eur Arch Otorhinolaryngol. 2020 Feb;277(2):611-621. doi: 10.1007/s00405-019-05732-x. Epub 2019 Dec 2. Eur Arch Otorhinolaryngol. 2020. PMID: 31792655 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical