AhR Activation Leads to Massive Mobilization of Myeloid-Derived Suppressor Cells with Immunosuppressive Activity through Regulation of CXCR2 and MicroRNA miR-150-5p and miR-543-3p That Target Anti-Inflammatory Genes - PubMed (original) (raw)
AhR Activation Leads to Massive Mobilization of Myeloid-Derived Suppressor Cells with Immunosuppressive Activity through Regulation of CXCR2 and MicroRNA miR-150-5p and miR-543-3p That Target Anti-Inflammatory Genes
Wurood Hantoosh Neamah et al. J Immunol. 2019.
Abstract
The compound 2,3,7,8-tetrachlorodibenzo-_p_-dioxin (TCDD), an environmental contaminant, is a potent ligand for aryl hydrocarbon receptor (AhR). In the current study, we made an exciting observation that naive C57BL/6 mice that were exposed i.p. to TCDD showed massive mobilization of myeloid-derived suppressor cells (MDSCs) in the peritoneal cavity. These MDSCs were highly immunosuppressive and attenuated Con A-induced hepatitis upon adoptive transfer. TCDD administration in naive mice also led to induction of several chemokines and cytokines in the peritoneal cavity and serum (CCL2, CCL3, CCL4, CCL11, CXCL1, CXCL2, CXCL5, CXCL9, G-CSF, GM-CSF, VEGF, and M-CSF) and chemokine receptors on MDSCs (CCR1, CCR5, and CXCR2). Treatment with CXCR2 or AhR antagonist in mice led to marked reduction in TCDD-induced MDSCs. TCDD-induced MDSCs had high mitochondrial respiration and glycolytic rate and exhibited differential microRNA (miRNA) expression profile. Specifically, there was significant downregulation of miR-150-5p and miR-543-3p. These two miRNAs targeted and enhanced anti-inflammatory and MDSC-regulatory genes, including IL-10, PIM1, ARG2, STAT3, CCL11 and its receptors CCR3 and CCR5 as well as CXCR2. The role of miRs in MDSC activation was confirmed by transfection studies. Together, the current study demonstrates that activation of AhR in naive mice triggers robust mobilization of MDSCs through induction of chemokines and their receptors and MDSC activation through regulation of miRNA expression. AhR ligands include diverse compounds from environmental toxicants, such as TCDD, that are carcinogenic to dietary indoles that are anti-inflammatory. Our studies provide new insights on how such ligands may regulate health and disease through induction of MDSCs.
Copyright © 2019 by The American Association of Immunologists, Inc.
Figures
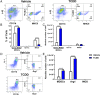
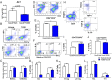
FIGURE 1.
TCDD induces MDSCs in naive mice. Naive C57BL/6 mice were injected with TCDD (n = 6) or vehicle (n = 6) i.p. and at various days posttreatment, and cells from the peritoneal cavity were harvested and analyzed for MDSCs. (A) Representative plots from FlowJo software analysis of flow cytometry data showing induced MDSC in percentages following 1, 5, or 10 μg/kg TCDD administration when compared with vehicle. Cells were harvested on day 3. (B) Total number of MDSCs per mouse expressed as mean ± SEM, based on (A) description. (C) Representative dot plots showing M-MDSCs (CD11b+Ly-6G−Ly-6Chi) and G-MDSCs (CD11b+Ly-6G+Ly-6Clow) percentages following administration of 1, 5, or 10 μg/kg TCDD when compared with vehicle. (D) Total number of M-MDSCs and G-MDSCs per mouse expressed as mean ± SEM. (E) Time course of induction of MDSCs by 5 μg/kg TCDD. (F) Based on data in (E), total number of MDSCs per mouse at different days expressed as mean ± SEM. Statistical analysis was performed using one-way ANOVA with Tukey post hoc multiple comparisons test for (B) and (D). For (F), a multiple t test was performed using Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: *p < 0.05, ***p < 0.001, ****p < 0.0001.
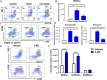
FIGURE 2.
MDSCs and MDSC subset characterization. Peritoneal exudate cells were harvested from vehicle- (n = 6) and TCDD-treated (n = 6) mice and stained for various markers. (A) Cells from vehicle- or TCDD-treated mice were gated on those that expressed CD11b+Gr-1+, and this population was further tested for MHC-II and CD11c. (B) Percentage of cells as shown in (A) that are negative or positive for MHC-II and CD11c markers. (C) Absolute number of CD11b+Gr-1+ cells per mouse that are negative or positive for MHC-II and CD11c markers. (D) Percentage of Arg1 and INOS expression in CD11b+Gr-1+ cells in vehicle and TCDD treatment groups. (E) Absolute number of CD11b+Gr-1+ cells and CD11b+Gr-1+ cells that express Arg1 and INOS. Vertical bars represent mean ± SEM. For bar graphs depicted, the multiple t test was performed using Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: ****p > 0.0001.
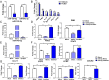
FIGURE 3.
AhR antagonist CH223191 treatment decreases TCDD-mediated MDSC induction. Naive C57BL/6 mice were injected with TCDD (10 μg/kg) i.p. as described in Fig. 1 legend. These mice were injected i.p with 10 mg/kg of AhR antagonist (CH223191, CH) 1 d before TCDD injection. Peritoneal exudates were collected on day 3 and stained for MDSCs. (A) Representative flow cytometric analysis showing MDSC percentages after treatment with AhR antagonist. (B) Total number of MDSCs per mouse expressed as mean ± SEM following treatment with AhR antagonist, based on description in (A). (C) Representative flow cytometric analysis showing percentages of MDSC subsets after treatment with AhR antagonist (CH). (D) Total number of MDSC subsets per mouse expressed as mean ± SEM following treatment with AhR antagonist. (E) Representative flow cytometric analysis showing percentages of MDSC (top panel) and MDSC subsets (bottom panel) after treatment with AhR agonist (3-MC). (F) Total number of MDSC subsets per mouse expressed as mean ± SEM following treatment with AhR agonist (3-MC). Statistical analysis was performed using one-way ANOVA with Tukey post hoc multiple comparisons test to determine significance for (B) and (D) between vehicle (n = 7) and TCDD (n = 7). For (F), significance between vehicle (n = 6) and 3-MC (n = 6) was determined using a multiple t test with Holm–Sidak method multiple comparisons test. Data are representative of at least two independent experiments with reproducible results. Significance was designated as follows: **p < 0.01, ***p < 0.001, ****p < 0.0001.
FIGURE 4.
Identifying the source of TCDD-induced MDSCs. Naive C57BL/6 mice were injected with TCDD (10 μg/kg) i.p. as described in Fig. 1 legend. (A) Representative flow cytometric analysis showing the percentage of MDSCs in BM 16 h after TCDD treatment when compared with 0 h (top panel). MDSCs percentage in peritoneal cavity (PC) after 16 h compared with 0 h (bottom panel). (B) Total number of MDSCs per mouse expressed as mean ± SEM, based on description in (A), where n = 5 for each experimental condition. (C) Measurement of chemokines from the peritoneal exudates of vehicle (n = 4) and TCDD (n = 4) mice with data expressed as mean ± SEM. (D) Detection of chemokines in the serum from vehicle- (n = 4) and TCDD-treated (n = 4) mice with data expressed as mean ± SEM. (E) Q-PCR validation of CCR1, CCR5, and CXCR2 expression in vehicle- (n = 4) and TCDD-treated (n = 4) mice, with data expressed as mean ± SEM. (F) Flow cytometric analysis of MDSC percentage (top panel) and absolute numbers (bottom panel) following treatment with vehicle (n = 5), TCDD (n = 5), or TCDD and CXCR2 antagonist (n = 3). (G) Representative plots of BrdU labeling and anti-Ki67 staining at 48 h after TCDD treatment. The left panel shows staining for MDSCs and right panel shows staining for BrdU and Ki67 on gated MDSCs. Vertical bars represent mean ± SEM. For (B)–(D), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. For (F), significance was determined using one-way ANOVA and Tukey multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
FIGURE 5.
TCDD-induced MDSCs suppress T cell proliferation and exhibit different metabolic profile. (A–C) TCDD-induced–purified PC-MDSCs or M-MDSCs and G-MDSCs were incubated with spleen cells activated with Con A at different ratios, and T cell proliferation was assessed by [3H]-thymidine incorporation assay. Data are depicted as mean ± SEM of triplicate cultures (n = 3) shown as counts per minute (CPM). (D and E) OCR and glycolytic PER in TCDD-induced PC-MDSCs (n = 3) compared with vehicle-induced PC-MDSCs (n = 3). (F) ATP production rate in the experimental groups including MDSCs from vehicle- (n = 3) or TCDD-treated mice (n = 3). For (A)–(C), one-way ANOVA with Tukey multiple comparisons test was used to determine significance. For (D) and (E), two-way ANOVA with Tukey multiple comparisons test was used to determine significance. For (E), significance, ****p < 0.0001, was found from 0 to 40 min time points and is illustrated with a bar across these six data points. For (F), a multiple t test with Holm–Sidak method of multiple comparison was used to determine significance. Data are representative of at least two independent experiments with reproducible results. Significance was defined as follows: p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
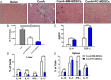
FIGURE 6.
TCDD-induced MDSCs protect from Con A–induced liver damage and inflammation in vivo following adoptive transfer. C57BL/6 mice were injected i.v. with Con A (12.5 mg/kg), and these mice received the injection 1 h before an adoptive transfer of 5 million purified PC-MDSCs or BM-MDSCs from TCDD-treated mice. Mice were sacrificed after 48 h posttreatment for further analysis to include the following groups: naive (n = 3), Con A (n = 3), Con A plus BM-MDSCs (n = 3), Con A plus PC-MDSCs (n = 3). (A) H&E stain of liver tissue at original magnification ×20. (B) Measurement of ALT in sera with significance determined by using ANOVA and Tukey multiple comparisons test. Vertical error bars represent mean ± SEM. (C) Measurement of TGF-β and IL-4 level in sera. (D and E) Percentages of cells expressing various cytokines determined by flow cytometry in spleen (D) and liver (E). For (C)–(E), one-way ANOVA with Tukey multiple comparisons test was used to determine significance, with vertical error bars representing mean ± SEM. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
FIGURE 7.
TCDD treatment attenuates Con A–induced hepatitis and associated inflammation. Con A was used to induce hepatitis as described in Fig. 6 legend. These mice received TCDD (10 μg/kg) by i.p. route 1 h before Con A injection followed by analysis of spleens and liver for inflammation. (A) ALT level in serum of hepatitis-induced mice treated with vehicle (n = 5) or TCDD (n = 5). (B and C) Percentage and total numbers of MDSCs and G-MDSCs in the spleens of two groups, respectively. (D and E) Percentage of CD3+CD4+ cells in the spleens, respectively. (F and G) Percentages of Th1 (CD3+CD4+IFNγ+) and Th2 (CD3+CD4+IL-4+) cells in splenocytes. (H–L) Percentage of Tregs and Treg subsets from spleens of experimental groups. Vertical error bars represent mean ± SEM. For (A), (I), and (J), significance was determined using a parametric unpaired two-tailed t test with Welch corrections. For (C), (G), (K), and (L), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
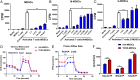
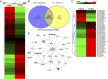
FIGURE 8.
TCDD-mediated alterations in miRNA expression in MDSCs. Naive mice were treated with TCDD as described in Fig. 1 legend. The PC-MDSCs were analyzed for miRNA expression between vehicle- and TCDD-treated mice, with MDSCs pooled from five different mice per experimental group. (A) Heat map of miRNAs expression in MDSCs from vehicle and TCDD groups showing more than 3000 miRNAs. (B) Venn diagram showing miRNA that are upregulated or downregulated in TCDD group when compared with vehicle controls. (C) Heat map of upregulated and downregulated (>2-fold) miRNAs in vehicle versus TCDD groups. (D) IPA was used to determine interaction between upregulated and downregulated miRNAs and targeted genes. Data are representative of one independent experiment.
FIGURE 9.
Q-PCR analysis of miRNA-150-5p and miRNA-543-3p and specific targeted genes. MDSCs were isolated as described in Fig. 1 legend in vehicle (n = 5) or TCDD (n = 5) mice. (A) Expression of mir-150-5p and mir-543-3p in TCDD-induced MDSCs when compared with vehicle. (B) Expression of targeted genes IL-10, PIM1, ARG2, STAT3, CCL11, CCR3, and CCR5 in TCDD-induced MDSCs when compared with vehicle. (C) Induction of mir-150-5p expression with mimic compared with mock and inhibitor. (D) Induction of IL-10 and PIM1 expression with inhibitor of mir-150-5p compared with mimic of mir-150-5p. (E) Expression of mir-543-3p with mimic compared with mock and inhibitor. (F and G) Expression of ARG2, STAT3, CCL11, CCR3, CCR5, and CXCR2 in the presence of inhibitor of mir-543-3p compared with mimic of mir-543-3p. For (A) and (B), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. For (C)–(G), using triplicate (n = 3) wells for each experimental condition, one-way ANOVA with Tukey multiple comparisons test was used to determine significance. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Similar articles
- AhR Activation Leads to Alterations in the Gut Microbiome with Consequent Effect on Induction of Myeloid Derived Suppressor Cells in a CXCR2-Dependent Manner.
Neamah WH, Busbee PB, Alghetaa H, Abdulla OA, Nagarkatti M, Nagarkatti P. Neamah WH, et al. Int J Mol Sci. 2020 Dec 17;21(24):9613. doi: 10.3390/ijms21249613. Int J Mol Sci. 2020. PMID: 33348596 Free PMC article. - Resveratrol Attenuates 2,3,7,8-Tetrachlorodibenzo-p-dioxin-Mediated Induction of Myeloid-Derived Suppressor Cells (MDSC) and Their Functions.
Neamah WH, Rutkovsky A, Abdullah O, Wilson K, Bloomquist R, Nagarkatti P, Nagarkatti M. Neamah WH, et al. Nutrients. 2023 Nov 3;15(21):4667. doi: 10.3390/nu15214667. Nutrients. 2023. PMID: 37960320 Free PMC article. - AhR Ligands Differentially Regulate miRNA-132 Which Targets HMGB1 and to Control the Differentiation of Tregs and Th-17 Cells During Delayed-Type Hypersensitivity Response.
Abdulla OA, Neamah W, Sultan M, Chatterjee S, Singh N, Nagarkatti M, Nagarkatti P. Abdulla OA, et al. Front Immunol. 2021 Feb 19;12:635903. doi: 10.3389/fimmu.2021.635903. eCollection 2021. Front Immunol. 2021. PMID: 33679792 Free PMC article. - From Suppressor T cells to Regulatory T cells: How the Journey That Began with the Discovery of the Toxic Effects of TCDD Led to Better Understanding of the Role of AhR in Immunoregulation.
Prasad Singh N, Nagarkatti M, Nagarkatti P. Prasad Singh N, et al. Int J Mol Sci. 2020 Oct 22;21(21):7849. doi: 10.3390/ijms21217849. Int J Mol Sci. 2020. PMID: 33105907 Free PMC article. Review. - Correlation between MDSC and Immune Tolerance in Transplantation: Cytokines, Pathways and Cell-cell Interaction.
Yang T, Li J, Li R, Yang C, Zhang W, Qiu Y, Yang C, Rong R. Yang T, et al. Curr Gene Ther. 2019;19(2):81-92. doi: 10.2174/1566523219666190618093707. Curr Gene Ther. 2019. PMID: 31237207 Review.
Cited by
- Environmental exposure and the role of AhR in the tumor microenvironment of breast cancer.
Sweeney C, Lazennec G, Vogel CFA. Sweeney C, et al. Front Pharmacol. 2022 Dec 15;13:1095289. doi: 10.3389/fphar.2022.1095289. eCollection 2022. Front Pharmacol. 2022. PMID: 36588678 Free PMC article. Review. - 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces multigenerational alterations in the expression of microRNA in the thymus through epigenetic modifications.
Singh NP, Yang X, Bam M, Nagarkatti M, Nagarkatti P. Singh NP, et al. PNAS Nexus. 2022 Dec 9;2(1):pgac290. doi: 10.1093/pnasnexus/pgac290. eCollection 2023 Jan. PNAS Nexus. 2022. PMID: 36712935 Free PMC article. - Amino Acid Transport and Metabolism in Myeloid Function.
Halaby MJ, McGaha TL. Halaby MJ, et al. Front Immunol. 2021 Aug 12;12:695238. doi: 10.3389/fimmu.2021.695238. eCollection 2021. Front Immunol. 2021. PMID: 34456909 Free PMC article. Review. - The aryl hydrocarbon receptor suppresses immunity to oral squamous cell carcinoma through immune checkpoint regulation.
Kenison JE, Wang Z, Yang K, Snyder M, Quintana FJ, Sherr DH. Kenison JE, et al. Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2012692118. doi: 10.1073/pnas.2012692118. Proc Natl Acad Sci U S A. 2021. PMID: 33941684 Free PMC article. - Effect of TCDD exposure in adult female and male mice on the expression of miRNA in the ovaries and testes and associated reproductive functions.
Hall A, Mattison D, Singh N, Chatzistamou I, Zhang J, Nagarkatti M, Nagarkatti P. Hall A, et al. Front Toxicol. 2023 Oct 3;5:1268293. doi: 10.3389/ftox.2023.1268293. eCollection 2023. Front Toxicol. 2023. PMID: 37854252 Free PMC article.
References
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. 1996. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140: 173–179. - PubMed
- McIntosh B. E., Hogenesch J. B., Bradfield C. A. 2010. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72: 625–645. - PubMed
- Schmidt J. V., Bradfield C. A. 1996. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12: 55–89. - PubMed
- Fisher M. T., Nagarkatti M., Nagarkatti P. S. 2004. Combined screening of thymocytes using apoptosis-specific cDNA array and promoter analysis yields novel gene targets mediating TCDD-induced toxicity. Toxicol. Sci. 78: 116–124. - PubMed
- Camacho I. A., Singh N., Hegde V. L., Nagarkatti M., Nagarkatti P. S. 2005. Treatment of mice with 2,3,7,8-tetrachlorodibenzo-p-dioxin leads to aryl hydrocarbon receptor-dependent nuclear translocation of NF-kappaB and expression of Fas ligand in thymic stromal cells and consequent apoptosis in T cells. J. Immunol. 175: 90–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI129788/AI/NIAID NIH HHS/United States
- R01 ES019313/ES/NIEHS NIH HHS/United States
- P20 GM103641/GM/NIGMS NIH HHS/United States
- R01 AI123947/AI/NIAID NIH HHS/United States
- R01 AT006888/AT/NCCIH NIH HHS/United States
- P01 AT003961/AT/NCCIH NIH HHS/United States
- R01 MH094755/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous