HH-suite3 for fast remote homology detection and deep protein annotation - PubMed (original) (raw)
HH-suite3 for fast remote homology detection and deep protein annotation
Martin Steinegger et al. BMC Bioinformatics. 2019.
Abstract
Background: HH-suite is a widely used open source software suite for sensitive sequence similarity searches and protein fold recognition. It is based on pairwise alignment of profile Hidden Markov models (HMMs), which represent multiple sequence alignments of homologous proteins.
Results: We developed a single-instruction multiple-data (SIMD) vectorized implementation of the Viterbi algorithm for profile HMM alignment and introduced various other speed-ups. These accelerated the search methods HHsearch by a factor 4 and HHblits by a factor 2 over the previous version 2.0.16. HHblits3 is ∼10× faster than PSI-BLAST and ∼20× faster than HMMER3. Jobs to perform HHsearch and HHblits searches with many query profile HMMs can be parallelized over cores and over cluster servers using OpenMP and message passing interface (MPI). The free, open-source, GPLv3-licensed software is available at https://github.com/soedinglab/hh-suite .
Conclusion: The added functionalities and increased speed of HHsearch and HHblits should facilitate their use in large-scale protein structure and function prediction, e.g. in metagenomics and genomics projects.
Keywords: Algorithm; Functional annotation; Homology detection; Profile HMM; Protein alignment; SIMD; Sequence search.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Fig. 1
HMM-HMM alignment of query and target. The alignment is represented as red path through both HMMs. The corresponding pair state sequence is MM, MM, MI, MM, MM, DG, MM
Fig. 2
SIMD parallelization over target profile HMMs. Batches of 4 or 8 database profile HMMs are aligned together by the vectorized Viterbi algorithm. Each cell (i,j) in the dynamic programming matrix is processed in parallel for 4 or 8 target HMMs
Fig. 3
The layout of the log transition probabilities (top) and emission probabilities (bottom) in memory for single-instruction single data (SISD) and SIMD algorithms. For the SIMD algorithm, 4 (using SSE2) or 8 (using AVX 2) target profile HMMs (t1 – t4) are stored together in interleaved fashion: the 4 or 8 transition or emission values at position i in these HMMs are stored consecutively (indicated by the same color). In this way, a single cache line read of 64 bytes can fill four SSE2 or two AVX2 SIMD registers with 4 or 8 values each
Fig. 4
Two approaches to reduce the memory requirement for the DP score matrices from O(L q L t) to O(L t), where L q and L t are lengths of the query and target profile, respectively. (Top) One vector holds the scores of the previous row, _S_XY(_i_−1,·), for pair state XY ∈{MM, MI, IM, GD and DG}, and the other holds the scores of the current row, _S_XY(i,·) for pair state XY ∈{MM, MI, IM, GD and DG}. Vector pointers are swapped after each row has been processed. (Bottom) A single vector per pair state XY holds the scores of the current row up to _j_−1 and of the previous row for j to L t. The second approach is somewhat faster and was chosen for HH-suite3
Fig. 5
Predecessor pair states for backtracing the Viterbi alignments are stored in a single byte of the backtrace matrix in HH-suite3 to reduce memory requirements. The bits 0 to 2 (blue) are used to store the predecessor state to the MM state, bits 3 to 6 store the predecessor of GD, IM, DG and MI pair states. The last bit denotes cells that are not allowed to be part of the suboptimal alignment because they are near to a cell that was part of a better-scoring alignment
Fig. 6
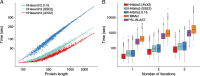
Speed comparisons. a runtime versus query profile length for 1644 searches with profile HMMs randomly sampled from UniProt. These queries were searched against the PDB70 database containing 35 000 profile HMMs of average length 234. The average speedup over HHsearch 2.0.16 is 3.2-fold for SSE2- vectorized HHsearch and 4.2-fold for AVX2-vectorized HHsearch. b Box plot for the distribution of total runtimes (in logarithmic scale) for one, two, or three search iterations using the 1644 profile HMMs as queries. PSI-BLAST and HHMER3 searches were done against the UniProt database (version 2015_06) containing 49 293 307 sequences. HHblits searches against the uniprot20 database, a clustered version of UniProt containing profile HMMs for each of its 7 313 957 sequence clusters. Colored numbers: speed-up factors relative to HMMER3
Fig. 7
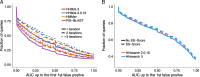
Sensitivity of sequence search tools. a We searched with 6616 SCOP20 domain sequences through the UniProt plus SCOP20 database using one to three search iterations. The sensitivity to detect homologous sequences is measured by cumulative distribution of the Area Under the Curve 1 (AUC1), the fraction of true positives ranked better than the first false positive match. True positive matches are defined as being from the same SCOP superfamily [25], false positives have different SCOP folds, excepting known cases of inter-fold homologies. b Sensitivity of HHsearch with and without scoring secondary structure similarity, measured by the cumulative distribution of AUC1 for a comparison of 6616 profile HMMs built from SCOP20 domain sequences. Query HMMs include predicted secondary structure, target HMMs include actual secondary structure annotated by DSSP. True and false positives are defined as in A
Similar articles
- Protein homology detection by HMM-HMM comparison.
Söding J. Söding J. Bioinformatics. 2005 Apr 1;21(7):951-60. doi: 10.1093/bioinformatics/bti125. Epub 2004 Nov 5. Bioinformatics. 2005. PMID: 15531603 - MMseqs software suite for fast and deep clustering and searching of large protein sequence sets.
Hauser M, Steinegger M, Söding J. Hauser M, et al. Bioinformatics. 2016 May 1;32(9):1323-30. doi: 10.1093/bioinformatics/btw006. Epub 2016 Jan 6. Bioinformatics. 2016. PMID: 26743509 - Accelerated Profile HMM Searches.
Eddy SR. Eddy SR. PLoS Comput Biol. 2011 Oct;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. Epub 2011 Oct 20. PLoS Comput Biol. 2011. PMID: 22039361 Free PMC article. - Profile hidden Markov models.
Eddy SR. Eddy SR. Bioinformatics. 1998;14(9):755-63. doi: 10.1093/bioinformatics/14.9.755. Bioinformatics. 1998. PMID: 9918945 Review. - Five hierarchical levels of sequence-structure correlation in proteins.
Bystroff C, Shao Y, Yuan X. Bystroff C, et al. Appl Bioinformatics. 2004;3(2-3):97-104. doi: 10.2165/00822942-200403020-00004. Appl Bioinformatics. 2004. PMID: 15693735 Review.
Cited by
- Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Marabini R, Condezo GN, Krupovic M, Menéndez-Conejero R, Gómez-Blanco J, San Martín C. Marabini R, et al. Sci Adv. 2021 Mar 31;7(14):eabe6008. doi: 10.1126/sciadv.abe6008. Print 2021 Mar. Sci Adv. 2021. PMID: 33789897 Free PMC article. - Comparative Computational Analysis of Spike Protein Structural Stability in SARS-CoV-2 Omicron Subvariants.
Balupuri A, Kim JM, Choi KE, No JS, Kim IH, Rhee JE, Kim EJ, Kang NS. Balupuri A, et al. Int J Mol Sci. 2023 Nov 8;24(22):16069. doi: 10.3390/ijms242216069. Int J Mol Sci. 2023. PMID: 38003257 Free PMC article. - Analysis of Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis Revealed Novel Groups of Filamentous Bacteriophages.
Evseev P, Bocharova J, Shagin D, Chebotar I. Evseev P, et al. Viruses. 2023 Nov 5;15(11):2215. doi: 10.3390/v15112215. Viruses. 2023. PMID: 38005892 Free PMC article. - Antibacterial properties and urease suppression ability of Lactobacillus inhibit the development of infectious urinary stones caused by Proteus mirabilis.
Szczerbiec D, Bednarska-Szczepaniak K, Torzewska A. Szczerbiec D, et al. Sci Rep. 2024 Jan 10;14(1):943. doi: 10.1038/s41598-024-51323-0. Sci Rep. 2024. PMID: 38200115 Free PMC article. - Antibacterial T6SS effectors with a VRR-Nuc domain are structure-specific nucleases.
Hespanhol JT, Sanchez-Limache DE, Nicastro GG, Mead L, Llontop EE, Chagas-Santos G, Farah CS, de Souza RF, Galhardo RDS, Lovering AL, Bayer-Santos E. Hespanhol JT, et al. Elife. 2022 Oct 13;11:e82437. doi: 10.7554/eLife.82437. Elife. 2022. PMID: 36226828 Free PMC article.
References
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23(1):205–11. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials