Genome sequence of Kobresia littledalei, the first chromosome-level genome in the family Cyperaceae - PubMed (original) (raw)
doi: 10.1038/s41597-020-0518-3.
Wei Wei # 1 2, Hailing Zi # 3, Magaweng Bai 1 2, Yunfei Liu 1 2, Dan Gao 3, Dengqunpei Tu 1 2, Yuhong Bao 1 2, Li Wang 1 2, Shaofeng Chen 1 2, Xing Zhao 4, Guangpeng Qu 5 6
Affiliations
- PMID: 32528014
- PMCID: PMC7289886
- DOI: 10.1038/s41597-020-0518-3
Genome sequence of Kobresia littledalei, the first chromosome-level genome in the family Cyperaceae
Muyou Can et al. Sci Data. 2020.
Abstract
Kobresia plants are important forage resources in the Qinghai-Tibet Plateau and are essential in maintaining the ecological balance of grasslands. Therefore, it is beneficial to obtain Kobresia genome resources and study the adaptive characteristics of Kobresia plants in the Qinghai-Tibetan Plateau. We assembled the genome of Kobresia littledalei C. B. Clarke, which was about 373.85 Mb in size. 96.82% of the bases were attached to 29 pseudo-chromosomes, combining PacBio, Illumina and Hi-C sequencing data. Additional investigation of the annotation identified 23,136 protein-coding genes. 98.95% of these were functionally annotated. According to phylogenetic analysis, K. littledalei in Cyperaceae separated from Poaceae about 97.6 million years ago after separating from Ananas comosus in Bromeliaceae about 114.3mya. For K. littledalei, we identified a high-quality genome at the chromosome level. This is the first time a reference genome has been established for a species of Cyperaceae. This genome will help additional studies focusing on the processes of plant adaptation to environments with high altitude and cold weather.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
A representative individual of Kobresia littledalei.
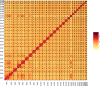
Fig. 2
Heat map of chromatin contact matrices generated by aligning a Hi-C dataset to the Kobresia littledalei genome. The frequency of interactions was calculated using a window size of 500 K.
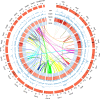
Fig. 3
Features of the Kobresia littledalei genome.
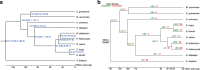
Fig. 4
The phylogenetic relationships and divergence times of commelinid plants, and contraction and expansion of gene families. (a) The phylogenetic relationships and divergence times of commelinid plants. Phylogenetic reconstructions using concatenation of 1,077 genes and the maximum likelihood (ML) method with A. thaliana as the distant outgroup. Divergence times were estimated using the ‘mcmctree’ program incorporated in the PAML package. (b) Contraction and expansion of gene families. Numbers in green represent expanded families on this clade, and numbers in red represent contracted families on this clade.
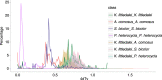
Fig. 5
Whole genome duplication events in Kobresia littledalei and other Poaceae plants.
Similar articles
- Draft genomes assembly and annotation of Carex parvula and Carex kokanica reveals stress-specific genes.
Qu G, Bao Y, Liao Y, Liu C, Zi H, Bai M, Liu Y, Tu D, Wang L, Chen S, Zhou G, Can M. Qu G, et al. Sci Rep. 2022 Mar 23;12(1):4970. doi: 10.1038/s41598-022-08783-z. Sci Rep. 2022. PMID: 35322069 Free PMC article. - The chromosome-scale genome of Kobresia myosuroides sheds light on karyotype evolution and recent diversification of a dominant herb group on the Qinghai-Tibet Plateau.
Ning Y, Li Y, Dong SB, Yang HG, Li CY, Xiong B, Yang J, Hu YK, Mu XY, Xia XF. Ning Y, et al. DNA Res. 2023 Feb 1;30(1):dsac049. doi: 10.1093/dnares/dsac049. DNA Res. 2023. PMID: 36503982 Free PMC article. - Identification and validation of stable reference genes for RT-qPCR analyses of Kobresia littledalei seedlings.
Sun H, Li C, Li S, Ma J, Li S, Li X, Gao C, Yang R, Ma N, Yang J, Yang P, He X, Hu T. Sun H, et al. BMC Plant Biol. 2024 May 11;24(1):389. doi: 10.1186/s12870-024-04924-w. BMC Plant Biol. 2024. PMID: 38730341 Free PMC article. - Research Progress on Dormancy Mechanism and Germination Technology of Kobresia Seeds.
Wang N, Zhang Z, Xu W, Zhou H, Ning R. Wang N, et al. Plants (Basel). 2022 Nov 22;11(23):3192. doi: 10.3390/plants11233192. Plants (Basel). 2022. PMID: 36501232 Free PMC article. Review. - The Kobresia pygmaea ecosystem of the Tibetan highlands - Origin, functioning and degradation of the world's largest pastoral alpine ecosystem: Kobresia pastures of Tibet.
Miehe G, Schleuss PM, Seeber E, Babel W, Biermann T, Braendle M, Chen F, Coners H, Foken T, Gerken T, Graf HF, Guggenberger G, Hafner S, Holzapfel M, Ingrisch J, Kuzyakov Y, Lai Z, Lehnert L, Leuschner C, Li X, Liu J, Liu S, Ma Y, Miehe S, Mosbrugger V, Noltie HJ, Schmidt J, Spielvogel S, Unteregelsbacher S, Wang Y, Willinghöfer S, Xu X, Yang Y, Zhang S, Opgenoorth L, Wesche K. Miehe G, et al. Sci Total Environ. 2019 Jan 15;648:754-771. doi: 10.1016/j.scitotenv.2018.08.164. Epub 2018 Aug 14. Sci Total Environ. 2019. PMID: 30134213 Review.
Cited by
- Draft genomes assembly and annotation of Carex parvula and Carex kokanica reveals stress-specific genes.
Qu G, Bao Y, Liao Y, Liu C, Zi H, Bai M, Liu Y, Tu D, Wang L, Chen S, Zhou G, Can M. Qu G, et al. Sci Rep. 2022 Mar 23;12(1):4970. doi: 10.1038/s41598-022-08783-z. Sci Rep. 2022. PMID: 35322069 Free PMC article. - The genome assembly of Carex breviculmis provides evidence for its phylogenetic localization and environmental adaptation.
Yuan T, Gao X, Xiang N, Wei P, Zhang G. Yuan T, et al. Ann Bot. 2024 Aug 22;134(3):467-484. doi: 10.1093/aob/mcae085. Ann Bot. 2024. PMID: 38822911 - The chromosome-scale genome of Kobresia myosuroides sheds light on karyotype evolution and recent diversification of a dominant herb group on the Qinghai-Tibet Plateau.
Ning Y, Li Y, Dong SB, Yang HG, Li CY, Xiong B, Yang J, Hu YK, Mu XY, Xia XF. Ning Y, et al. DNA Res. 2023 Feb 1;30(1):dsac049. doi: 10.1093/dnares/dsac049. DNA Res. 2023. PMID: 36503982 Free PMC article. - Phylogenomic profiles of whole-genome duplications in Poaceae and landscape of differential duplicate retention and losses among major Poaceae lineages.
Zhang T, Huang W, Zhang L, Li DZ, Qi J, Ma H. Zhang T, et al. Nat Commun. 2024 Apr 17;15(1):3305. doi: 10.1038/s41467-024-47428-9. Nat Commun. 2024. PMID: 38632270 Free PMC article.
References
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–453. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous