Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae - PubMed (original) (raw)
Review
Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae
Tania Jordá et al. Genes (Basel). 2020.
Abstract
Ergosterol is an essential component of fungal cell membranes that determines the fluidity, permeability and activity of membrane-associated proteins. Ergosterol biosynthesis is a complex and highly energy-consuming pathway that involves the participation of many enzymes. Deficiencies in sterol biosynthesis cause pleiotropic defects that limit cellular proliferation and adaptation to stress. Thereby, fungal ergosterol levels are tightly controlled by the bioavailability of particular metabolites (e.g., sterols, oxygen and iron) and environmental conditions. The regulation of ergosterol synthesis is achieved by overlapping mechanisms that include transcriptional expression, feedback inhibition of enzymes and changes in their subcellular localization. In the budding yeast Saccharomyces cerevisiae, the sterol regulatory element (SRE)-binding proteins Upc2 and Ecm22, the heme-binding protein Hap1 and the repressor factors Rox1 and Mot3 coordinate ergosterol biosynthesis (ERG) gene expression. Here, we summarize the sterol biosynthesis, transport and detoxification systems of S. cerevisiae, as well as its adaptive response to sterol depletion, low oxygen, hyperosmotic stress and iron deficiency. Because of the large number of ERG genes and the crosstalk between different environmental signals and pathways, many aspects of ergosterol regulation are still unknown. The study of sterol metabolism and its regulation is highly relevant due to its wide applications in antifungal treatments, as well as in food and pharmaceutical industries.
Keywords: Saccharomyces cerevisiae; ergosterol; iron; oxygen; sterol biosynthesis; sterol regulation; yeast.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
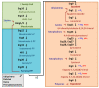
Figure 1
Ergosterol biosynthetic pathway in S. cerevisiae. The different color boxes represent the three modules into which the pathway can be divided: the green box is the mevalonate pathway, which occurs in the vacuole and mitochondria; the blue box consists of farnesyl pyrophosphate (farnesyl-PP) biosynthesis and is carried out in the vacuole; and the orange box contains the late pathway, which ends with ergosterol biosynthesis, and mainly takes place in the ER. In addition, farnesyl-PP is used in the synthesis of ubiquinone, dolichol, heme and prenylated proteins (gray box). Enzymes, intermediates, inhibitors and requirements of oxygen, heme and iron are indicated. Enzymes: Erg10, acetyl-CoA C-acetyltransferase; Erg13, HMG-CoA synthase; Hmg1/2, HMG-CoA reductase; Erg12, mevalonate kinase; Erg8, phosphomevalonate kinase; Mvd1/Erg19, mevalonate pyrophosphate decarboxylase; Idi1, Isopentenyl diphosphate Isomerase; Erg20, farnesyl pyrophosphate synthetase; Erg9, squalene synthase; Erg1, squalene epoxidase; Erg7, lanosterol synthase; Erg11 (Cyp51), lanosterol C-14 demethylase; Erg24, sterol C-14 reductase; Erg25, sterol C-4 methyloxydase; Erg26, sterol C-3 dehydrogenase (C4-decarboxylase); Erg27, sterol C-3 ketoreductase; Erg6, sterol C-24 methyltransferase; Erg2, sterol C-8 isomerase; Erg3, sterol C-5 desaturase; Erg5, sterol C-22 desaturase; Erg4, sterol C-24 reductase. Inhibitors: statins target Hmg1/2; allylamines inhibit Erg1; azoles inhibit Erg11; morpholines target Erg2 and, mainly, Erg24; polyenes bind ergosterol.
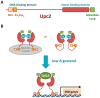
Figure 2
Structure and regulation of the S. cerevisiae Upc2 transcriptional factor. (A) Schematic representation of the primary structure of yeast Upc2 protein. NLS, nuclear localization signal. (B) Proposed model for the transcriptional regulation of Upc2. Under high-sterol conditions, Upc2 associates to ergosterol (Erg) and localizes to the cytosol. Although the mechanism is currently unknown, it has been proposed that Upc2 stays in the cytosol either because it interacts with a cytosolic protein or because its NLS is not available for nuclear import. Upon sterol depletion, ergosterol-free Upc2 undergoes a conformational change that allows its import to the nucleus, where it binds to ERG gene promoters to activate their transcription through the recruitment of a co-activator, probably the SAGA complex.
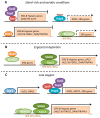
Figure 3
Transcriptional regulation of S. cerevisiae genes involved in ergosterol biosynthesis and uptake in response to low sterols or oxygen. (A) Under normal growth conditions, basal ERG gene expression is mostly maintained by Ecm22 and Hap1. Moreover, Mot3 and Rox1 (in a Tup1-Ssn6-dependent manner) inhibit the expression of hypoxic and ergosterol uptake genes. (B) Upon sterol depletion, Upc2 activates the transcription of ERG genes probably by recruiting the SAGA complex. (C) A drop in oxygen availability decreases heme levels, which lead to the Tup1-Ssn6 and Set4 recruitment by Hap1 to repress the expression of some ERG genes, ROX1 and MOT3. The decrease in Rox1 and Mot3 levels increases the expression of ERG, hypoxic and sterol uptake genes in a Upc2-Ecm22-dependent manner. Finally, Sut1 activates the transcription of AUS1 and DAN1, but not PDR11.
Similar articles
- The yeast anaerobic response element AR1b regulates aerobic antifungal drug-dependent sterol gene expression.
Gallo-Ebert C, Donigan M, Liu HY, Pascual F, Manners M, Pandya D, Swanson R, Gallagher D, Chen W, Carman GM, Nickels JT Jr. Gallo-Ebert C, et al. J Biol Chem. 2013 Dec 6;288(49):35466-77. doi: 10.1074/jbc.M113.526087. Epub 2013 Oct 25. J Biol Chem. 2013. PMID: 24163365 Free PMC article. - Loss-of-Function ROX1 Mutations Suppress the Fluconazole Susceptibility of _upc2A_Δ Mutation in Candida glabrata, Implicating Additional Positive Regulators of Ergosterol Biosynthesis.
Ollinger TL, Vu B, Murante D, Parker JE, Simonicova L, Doorley L, Stamnes MA, Kelly SL, Rogers PD, Moye-Rowley WS, Krysan DJ. Ollinger TL, et al. mSphere. 2021 Dec 22;6(6):e0083021. doi: 10.1128/msphere.00830-21. Epub 2021 Dec 22. mSphere. 2021. PMID: 34935446 Free PMC article. - Transcriptional regulation of ergosterol biosynthesis genes in response to iron deficiency.
Jordá T, Barba-Aliaga M, Rozès N, Alepuz P, Martínez-Pastor MT, Puig S. Jordá T, et al. Environ Microbiol. 2022 Nov;24(11):5248-5260. doi: 10.1111/1462-2920.16157. Epub 2022 Aug 12. Environ Microbiol. 2022. PMID: 36382795 Free PMC article. - Ergosterol Turnover in Yeast: An Interplay between Biosynthesis and Transport.
Sokolov SS, Trushina NI, Severin FF, Knorre DA. Sokolov SS, et al. Biochemistry (Mosc). 2019 Apr;84(4):346-357. doi: 10.1134/S0006297919040023. Biochemistry (Mosc). 2019. PMID: 31228926 Review. - The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn.
Lv QZ, Yan L, Jiang YY. Lv QZ, et al. Virulence. 2016 Aug 17;7(6):649-59. doi: 10.1080/21505594.2016.1188236. Epub 2016 May 24. Virulence. 2016. PMID: 27221657 Free PMC article. Review.
Cited by
- Exofacial membrane composition and lipid metabolism regulates plasma membrane P4-ATPase substrate specificity.
Jain BK, Roland BP, Graham TR. Jain BK, et al. J Biol Chem. 2020 Dec 25;295(52):17997-18009. doi: 10.1074/jbc.RA120.014794. Epub 2020 Oct 15. J Biol Chem. 2020. PMID: 33060204 Free PMC article. - Identification of Lipopeptide Iturin A Produced by Bacillus amyloliquefaciens NCPSJ7 and Its Antifungal Activities against Fusarium oxysporum f. sp. niveum.
Wang J, Qiu J, Yang X, Yang J, Zhao S, Zhou Q, Chen L. Wang J, et al. Foods. 2022 Sep 26;11(19):2996. doi: 10.3390/foods11192996. Foods. 2022. PMID: 36230072 Free PMC article. - In vitro and in vivo synergistic inhibition of Malassezia furfur targeting cell membranes by Rosa rugosa Thunb. and Coptidis Rhizoma extracts.
Li L, He Y, Zou Q, Chen W, Liu Y, He H, Zhang J. Li L, et al. Front Microbiol. 2024 Sep 11;15:1456240. doi: 10.3389/fmicb.2024.1456240. eCollection 2024. Front Microbiol. 2024. PMID: 39323889 Free PMC article. - Sexual Crossing, Chromosome-Level Genome Sequences, and Comparative Genomic Analyses for the Medicinal Mushroom Taiwanofungus Camphoratus (Syn. Antrodia Cinnamomea, Antrodia Camphorata).
Chen CL, Li WC, Chuang YC, Liu HC, Huang CH, Lo KY, Chen CY, Chang FM, Chang GA, Lin YL, Yang WD, Su CH, Yeh TM, Wang TF. Chen CL, et al. Microbiol Spectr. 2022 Feb 23;10(1):e0203221. doi: 10.1128/spectrum.02032-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196809 Free PMC article. - The emerging threat antifungal-resistant Candida tropicalis in humans, animals, and environment.
Lima R, Ribeiro FC, Colombo AL, de Almeida JN Jr. Lima R, et al. Front Fungal Biol. 2022 Aug 15;3:957021. doi: 10.3389/ffunb.2022.957021. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746212 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous