TDP-43 condensation properties specify its RNA-binding and regulatory repertoire - PubMed (original) (raw)
. 2021 Sep 2;184(18):4680-4696.e22.
doi: 10.1016/j.cell.2021.07.018. Epub 2021 Aug 10.
Anob M Chakrabarti 2, Flora C Y Lee 3, Bo Lim Lee 4, Aram G Amalietti 5, Hana M Odeh 4, Katie E Copley 6, Jack D Rubien 4, Bede Portz 4, Klara Kuret 7, Ina Huppertz 8, Frédérique Rau 3, Rickie Patani 3, Nicolas L Fawzi 9, James Shorter 6, Nicholas M Luscombe 10, Jernej Ule 11
Affiliations
- PMID: 34380047
- PMCID: PMC8445024
- DOI: 10.1016/j.cell.2021.07.018
TDP-43 condensation properties specify its RNA-binding and regulatory repertoire
Martina Hallegger et al. Cell. 2021.
Abstract
Mutations causing amyotrophic lateral sclerosis (ALS) often affect the condensation properties of RNA-binding proteins (RBPs). However, the role of RBP condensation in the specificity and function of protein-RNA complexes remains unclear. We created a series of TDP-43 C-terminal domain (CTD) variants that exhibited a gradient of low to high condensation propensity, as observed in vitro and by nuclear mobility and foci formation. Notably, a capacity for condensation was required for efficient TDP-43 assembly on subsets of RNA-binding regions, which contain unusually long clusters of motifs of characteristic types and density. These "binding-region condensates" are promoted by homomeric CTD-driven interactions and required for efficient regulation of a subset of bound transcripts, including autoregulation of TDP-43 mRNA. We establish that RBP condensation can occur in a binding-region-specific manner to selectively modulate transcriptome-wide RNA regulation, which has implications for remodeling RNA networks in the context of signaling, disease, and evolution.
Keywords: RNA granules; RNA-binding protein; TDP-43; alternative polyadenylation; amyotrophic lateral sclerosis; condensation; iCLIP; intrinsically disordered region; multivalency; phase separation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.S. is a consultant for Dewpoint Therapeutics, Maze Therapeutics, and Vivid Sciences. B.P. is an employee of Dewpoint Therapeutics.
Figures
Graphical abstract
Figure 1
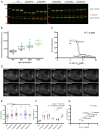
Deletions within the CR change the condensation behavior of TDP-43 in vitro (A) The domain map of TDP-43 includes an N-terminal domain (NTD), two RNA recognition motifs (RRM1 and RRM2), two intrinsically disordered regions (IDR1, 2) at the C terminus with an intervening conserved region (CR helix), and a nuclear localization signal (NLS; residues 82–92). The positions of the 5 deletion variants are shown. (B) Disorder confidence score, amino acid conservation (green, least conserved; red, most conserved position), and compositional biases of the C-terminal domain (STAR Methods). (C) Differential interference contrast (DIC) microscopy images of 10 μM WT MBP-TDP-43 and deletion variants show differences in droplet formation. The scale bar represents 10 μm. (D) Turbidity measurements of phase-separated WT MBP-TDP-43 and deletion variants. Mean (±SEM), n = 3, one-way ANOVA (∗∗p < 0.01 and ∗∗∗p < 0.001), significant against the WT. The dashed line indicates the WT absorbance value. (E) Csat (μM) of TDP-43 deletion constructs were determined by measuring the supernatant concentration after LLPS at 10 μM. Mean (±SEM), n = 3, one-way ANOVA shows significant difference against WT TDP-43 (∗∗∗p < 0.001). The dashed line indicates the Csat of the WT. (F) Phase diagram showing changes in the phase boundary of deletion variants. “Sparse, small droplets” refer to droplets as in 320del414 in (C). The experiment was repeated three times.
Figure S1
Deletions within the CR change the condensation behavior of TDP-43, related to Figure 1 A. Brightfield microscopy images of WT TDP-43-MBP and deletion variants (5μM) after addition of TEV protease show differences in droplet formation. The black bar represents 10 μm. B. Turbidity measurements of phase-separated WT TDP-43-MBP and deletion variants (5μM) after addition of TEV protease. Mean (±SEM), n = 2, one-way ANOVA (∗p < 0.05). Asterisks, significant relative to the WT. The dashed line indicates the absorbance value of WT. C. Phase diagram (left) shows changes in the phase boundary of deletion variants. Representative brightfield microscopy images are shown on the right. Scale bar, 10μm. The experiment was repeated two times. D. Brightfield microscopy images of WT TDP-43-MBP and deletion variants (10μM) after addition of TEV protease in the presence of 5ng/μl (upper panels) or 10ng/μl (lower panels) of total HeLa cell RNA show differences in droplet formation. The black bar represents 10 μm. E. Turbidity measurements of phase-separated WT TDP-43-MBP and deletion variants (10μM) after addition of TEV protease in the presence of 5ng/μl (left) or 10ng/μl (right) of total HeLa cell RNA. Mean (±SEM), n = 3-4, one-way ANOVA (∗∗p < 0.005; ∗∗∗p < 0.001). Asterisks, significant relative to the WT. The dashed line indicates the absorbance value of WT.
Figure S2
Deletions within the CR change the condensation behavior of TDP-43 in cells, related to Figure 2 A. Western blot analysis of expression level of the various dox-inducible constructs, compared to the endogenous TDP-43, as determined by the anti-TDP-43 antibody. B. Quantification of mean nuclear fluorescence levels of GFP-TDP-43 WT Flp-In cells induced with dox for 4, 8, 24 and 72hrs. n (cells) = 26, 27, 36, 29. C. Relationship between in vitro Csat (μM) of purified TDP-43 deletion constructs (Figure 1E) and quantification of foci counts per nucleus in confocal images of HEK293 Flp-In cell lines expressing the dox-inducible GFP-TDP-43 variants (Figure 2E). The sigmoidal curve (±95% confidence band) shown was fitted to the means of each datapoint. D. Representative image series and ROI from nucleoplasm FRAP experiments (Figure 2F) of GFP-TDP-43 WT cells. E. Mobile fraction of TDP-43, obtained from the plateau of the fitted exponential curve from FRAP data shown in Figure 2F. Mean ± 95%CI are shown for n (cells) = 34 for all cell lines. Significance was tested with Kruskal-Wallis test followed by Dunn’s Multiple Comparison Test. The p values reported are for the individual comparisons (∗p adj. < 0.05). F. Rate constant of FRAP from WT and deletion constructs with or without siRNA mediated knockdown of endogenous TDP-43 (siT). Mean ± 95%CI are shown for n (cells) = 12 for all conditions except n (cells) = 11 for 367del414 siT. Significance was tested with Two-way ANOVA (cell line: ∗∗∗∗p < 0.0001, siT: not significant p = 0.2249, interaction: not significant p = 0.3052). Rate constant was not found to be significantly different between all pairs of untreated versus si-TDP-43 conditions. G. Relationship between in vitro Csat (μM) of purified TDP-43 deletion constructs (Figure 1E) and FRAP rate constants assessing mobility of GFP-TDP-43 in HEK293 Flp-In cell lines (Figure 2G). The linear regression (±95% confidence band) shown was fitted to the means of each datapoint.
Figure 2
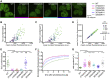
Deletions within the CR change the condensation behavior of TDP-43 in cells (A) Maximum z-projection images of HEK293 Flp-In cell lines expressing doxycycline (dox)-inducible GFP-TDP-43 variants. (B) Heterogeneous expression levels of GFP-TDP-43 WT were induced with a dox time course (4–72 h) in HEK293 Flp-In cells, n (cells) = 118. Foci counts in each segmented nucleus are plotted against the total nuclear fluorescence of individual GFP-TDP-43 WT cells. (C) As in (B), the fractional nuclear area occupied by the sum of all foci in each nucleus plotted against the mean nuclear fluorescence. (D) As in (B), the relationship between the mean foci fraction fluorescence intensity and mean nuclear fluorescence. (E) Quantification of foci counts in each segmented nucleus. Mean ± 95% confidence interval (CI) is shown. n (cells): WT = 21, 274del319 = 20, 316del346 = 22, 320del366 = 20, 367del414 = 23, 320del414 = 23. (F) FRAP experiments on HEK293 cell lines. The fluorescence recovery curve was obtained by bleaching a spot of predefined size in the nucleoplasm. Mean ± 95% CI is shown for 34 cells for all cell lines. (G) Rate constant of fluorescence recovery. Mean ± 95% CI is shown. n (cells) = 34 for all cell lines. Significance for (E) and (G) was tested with Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Reported adjusted (adj.) p values are for the individual comparisons (∗p adj. < 0.05, ∗∗∗p adj. < 0.001, and ∗∗∗∗p adj. < 0.0001).
Figure 3
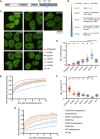
CR point mutants of TDP-43 have a gradient of in vivo condensation properties (A) Positions of the 5 point mutations in TDP-43 variants. (B) Mutations modify condensation on a gradient from perturbing (red/orange) to maintaining (green) or promoting (blue) the condensation capacity. (C) Maximum z-projection images of HEK293 Flp-In cell lines expressing the indicated dox-inducible GFP-TDP-43 variants. (D) Quantification of foci counts in each segmented nucleus of confocal z stacks. Mean ± 95% CI is shown for n (cells): 316del346 = 30, A326P = 32, M337P = 35, Q331K = 32, G294A = 32, G335A = 30, WT = 30. (E) FRAP experiments on GFP-TDP-43 CR mutant cell lines. The fluorescence recovery curve was obtained after bleaching a spot of predefined size in the nucleoplasm. Mean ± 95% CI is shown for n (cells): 316del346 = 37, A326P = 36, M337P = 36, Q331K = 36, G294A = 36, G335A = 36, WT = 48. (F) Rate constant of fluorescence recovery. Mean ± 95% CI is shown for the same number of cells as in (E). Significance for (D) and (F) was tested with Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Reported p values are for the individual comparisons (∗∗p adj. < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001). (G) As in (E) for FRAP analysis of GFP TDP-43 mobility in nucleoplasm versus nuclear regions centered on foci on 316del346, A326P, G335A, and WT GFP-TDP-43 cell lines. Mean ± 95% CI is shown for n (cells): 316del346 nucleoplasm = 8, 316del346 foci = 12, A326P nucleoplasm = 8, A326P foci = 12, G335A nucleoplasm = 8, G335A foci = 12, WT nucleoplasm = 8, WT foci = 10, 1 focus per independent cell.
Figure S3
CR point mutants of TDP-43 have a gradient of in vivo condensation properties, related to Figure 3 A. Mobile fraction obtained from the plateau of the fitted exponential curve from FRAP experiments shown in Figure 3E. Mean ± 95%CI are shown for n (cells): 316del346 = 37, A326P = 36, M337P = 36, Q331K = 36, G294A = 36, G335A = 36, WT = 48. Significance was tested with Kruskal-Wallis test followed by Dunn’s Multiple Comparison Test (∗∗p adj. < 0.01). B. Rate constant of fluorescence recovery from each of the 316del346, A326P, G335A and WT GFP-TDP-43 cell lines, in foci-centered regions or the surrounding nucleoplasm obtained from FRAP experiments shown in Figure 3G. Mean ± 95%CI are shown for n (cells): 316del346 nucleoplasmic = 8, 316del346 foci = 12, A326P nucleoplasmic = 8, A326P foci = 12, G335A nucleoplasmic = 8, G335A foci = 12, WT nucleoplasmic = 8, WT foci = 10, 1 focus per independent cell. Significance was tested with Two-way ANOVA (cell line: ∗∗∗∗p < 0.0001, nucleoplasm versus foci: ∗∗∗p = 0.0008, interaction: not significant p = 0.6656). C. As in Figure S2D, representative image series and ROI from focus-centered FRAP experiments (Figure 3G) of GFP-TDP-43 WT cells.
Figure 4
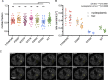
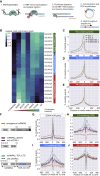
iCLIP reveals that condensation properties affect TDP-43 binding to specific RNA motifs (A) iCLIP involves UV-C crosslinking, cell lysis, RNA fragmentation by RNaseI, immunoprecipitation (IP) of crosslinked RBP-RNA complexes, and ligation to an infrared dye-labeled linker. After SDS-PAGE and transfer to a nitrocellulose membrane, RNA is released by proteinase K digestion, reverse transcribed, circularized, and PCR amplified. (B) The relative enrichment of the 20 6-mers that are most enriched across the “Mutants, low RNase experiment” iCLIP experiment (Table S1). The mean intronic motif enrichment of two replicates of each TDP-43 variant was normalized by the mean enrichment across all variants to define relative enrichment and plotted on the heatmap (log2 scale). The motifs were sorted based on the gradient of enrichment across TDP-43 variants and combined into three groups that are named according to the dominant sequence consensus: YG-containing [UG]n (green), YA-containing [UG]n (blue) or AA-containing [UG]n (red) (where Y indicates C or U). (C–E) Metaprofiles of YG-, YA-, or AA-containing [UG]n coverage around crosslink events in introns of replicates from the “RNase experiment" (Table S1). (F) hnRNPA2 domain map and design of its CTD deletion and TDP-43 fusion variants that were used for the “chimeraRBP-CLIP” experiment (Table S1). (G–J) Metaprofile of GGAA-type, YG-, YA-, or AA-containing [UG]n coverage around crosslink events in introns of replicates from the “chimeraRBP-CLIP” experiment, including samples with endogenous TDP-43 depletion (siTDP43).
Figure S4
iCLIP reveals that condensation properties affect TDP-43 binding to specific RNA motifs, related to Figure 4 A. In this representative iCLIP experiment, the RNA/TDP-43 complex was visualized by Li-Cor scanning of nitrocellulose membrane, which detects the infrared adaptor that is ligated to the protein-RNA complexes. Shown here are WT (MW 85kDa) and 316del346 construct (MW 81kDa). The shift of the RNA-TDP-43 complex to higher molecular weight - highlighted by the orange box - is caused by the cross-linked RNA and ligated adaptor. RNase concentration was at 0.4 (+, low), 2 units (++, high) per 1 mL of lysate at 1mg/ml protein concentration and libraries were produced from the two low-concentration replicates. B. The number of unique cDNAs mapping to each region of transcriptome is shown for each replicate for the experiment shown in Figure 4B, and compared to the iCLIP with the endogenous TDP-43. C. Metaprofile of YG-, YA- or AA-containing [UG]n coverage around crosslink events in 3′UTRs of replicates from the ‘RNase’ iCLIP experiment. D. Top: Maximum z-projection images of HEK293 Flp-In cell lines expressing the indicated dox-inducible GFP-TDP-43 WT, transfected with mCherry-hnRNPA2-delCTD or mCherry-hnRNPA2-TDP-CTD. Bottom: HEK293 Flp-In cell lines expressing dox inducible GFP-hnRNPA2-delCTD or GFP-hnRNPA2-TDP-CTD. E. As in (D) Boxplot showing quantification of GFP-TDP-43 WT foci count per nucleus upon transfection with the indicated mCherry-hnRNPA2 construct. Nuclear segmentation of images from coverslips transfected with mCherry-hnRNPA2 were blinded, then the mCherry channel was classified as low or high expression manually, two replicate experiments. n (cells): replicate 1: A2-delCTD low = 8, A2-delCTD high = 19, A2-TDP-CTD low = 10, A2-TDP-CTD high = 21; replicate 2: no plasmid control transfection = 18, A2-delCTD low = 9, A2-delCTD high = 13, A2-TDP-CTD low = 14, A2-TDP-CTD high = 10. Significance was tested with t test. F. Metaprofile of GGAA-type motif, and YG-, YA-, AA-containing [UG]n around crosslink events in 3′UTRs of replicates from the ‘chimeraRBP-CLIP′ iCLIP experiment, including samples with endogenous TDP-43 depleted (siTDP43).
Figure S5
Three RNA features define the condensation-binding relationships, related to Figure 5 A. Normalized gene expression for the genes with regulated PAS (see Figure 6A) in control and after WT or TDP-43 variant induction. B. The features of each binding region class, which contribute to their classification. The 36 classes of binding regions are ordered as in Figure 5, and the following features are shown: the number of regions in each class, the average cDNA count per replicate in the ‘HD’ experiment (Table S1), average length of regions in each class, the average density of all evaluated motifs, and the average coverage of YG- or AA-containing [UG]n motifs. C. A separate experiment was analyzed as explained in Figure 5A of TDP-43 mutant lines at a lower RNaseI concentration (0.2 units per 1 mL of lysate) resulting in longer RNA fragments. D. A separate experiment was analyzed as explained in Figure 5A, containing WT TDP-43 and 316del346 triplicate samples pre-treated either with 1,6- or 2,5-HD. C and D are linked to Table S3 containing quantification of cDNA counts from CLIP samples overlapping with the binding regions, together with their genomic coordinates, region, gene id and gene names and derived classifications in groups by length, density and base content. E. Maximum z-projection of confocal z stacks of dox-induced GFP-TDP-43 WT in HEK Flp-In cells after incubation with 8% 2,5-HD or 8% 1,6-HD for 5 minutes. F. Quantification of foci counts in each segmented nucleus from confocal z stacks. n (counted cells): 2,5-HD = 7, 1,6-HD = 5; segmented from n (fields of view): 2,5-HD = 2, 1,6-HD = 2. Significance was tested with a Welch Two Sample t test (∗p = 0.017). G. Turbidity measurements on pre-formed TDP-43-MBP condensates show a reduction in turbidity after addition of 1,6-HD compared to addition of buffer (n = 3, two-tailed t test, p < 0.0001). H. Alexa488-labeled TDP-43-MBP (1:200 labeled:unlabeled) was used to image TDP-43-MBP condensates after addition of buffer or 1,6-HD. Scale bar, 10 μm. I. Decrease in area fraction of TDP-43-MBP condensates upon addition of 1,6-HD compared to addition of buffer (n= 15 images, two-tailed t test, p < 0.0001). J. Decrease in TDP-43-MBP condensate size upon addition of 1,6-HD compared to addition of buffer (n = 15 images, two-tailed t test, p < 0.0001).
Figure 5
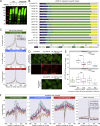
Three RNA features define the condensation-binding relationships (A) Binding regions were defined based on motifs proximal to crosslinks, and each region was allocated to one of 36 classes defined by the region length, motif density, and dominant motif type. The count of cDNAs falling into each class of binding regions from each iCLIP dataset was determined and normalized by the average cDNA count across all datasets within this experiment. Duplicate samples were obtained for each TDP-43 variant. Blue color represents depletion compared with the average, and gray represents enrichment. This is linked to Table S2 containing quantification of cDNA counts from CLIP samples overlapping with the binding regions together with their genomic coordinates, region, gene ID and gene names, and derived classifications in groups by length, density, and base content.
Figure 6
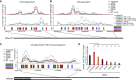
TDP-43 shows distinct condensation-dependent binding, and CR mutants have defects in autoregulation (A) Mapping of TDP-43 iCLIP data onto Malat1 non-coding RNA. Two replicates were summed, and iCLIP data were normalized and converted into smoothed lines using the rollmean function with a window size of 20 and mapped to a 250-nt-long regions on the ncRNA Malat1 (hg38 chr11:65501021-65501271:+) with CR-independent binding behavior. A crosslinking signal derived from the following two different iCLIP experiments is shown. Top panel: CR mutant TDP-43 variants. Center panel: hnRNPA2 constructs as described in Figure 4F. Bottom panel: the assigned binding regions colored according to their motif bias: YG-, YA-, and AA-containing [UG]n in green, blue, and red, respectively. (B) As in (A) for a CR-dependent and 1,6-HD-sensitive binding region on a 400-nt-long region of the ncRNA Malat1 (hg38 chr11:65504300-65504700:+). (C) As in (A) for a CR-dependent and 1,6-HD-sensitive region in the 3′ UTR of the endogenous TARDBP RNA (hg38 chr1:11023414-11023698:+). (D) Quantification of the western blot analysis of the endogenous TDP-43 levels after 2 days of induction of each of the GFP-TDP-43 variants; see the corresponding western blot in Figure S6B. Statistical significance of n = 3 was calculated using Student’s t test with ∗p < 0.05, ∗∗p < 0.01.
Figure S6
TDP-43 shows distinct condensation-dependent binding, and CR mutants have defects in autoregulation, related to Figure 6 A. Mapping of TDP-43 iCLIP data onto Neat1 ncRNA. Two replicates were summed and iCLIP data was normalized and converted into smoothed lines using rollmean with window size of 500 to two 22kB long regions on the ncRNA Neat1 with CR-dependent and -independent binding behavior. Crosslinking signal was derived from CR mutant TDP-43 variants. The bottom panel shows a motif-based binding site assignment where 300nt regions are colored according to their motif bias: YG-, YA-, AA-containing [UG]n in green and blue and red, respectively. B. Western blot analysis of expression level of the endogenous TDP-43 and GFP-TDP-43 variants after 2 days of induction with doxycycline (DOX), as determined by the anti-TDP-43 antibody. Data was compared to no DOX level of the endogenous TDP-43 protein and loading was normalized by alpha-tubulin as a loading control. C. Quantification of western blot analysis of the endogenous TDP-43 levels after two days of induction of each of the GFP-TDP-43 variants. Each sample replicate was normalized by alpha-tubulin as a loading control. D. Same as in (C) except, each sample replicate was normalized by GFP-TDP-43 variant expression.
Figure S7
Regulation of a subset of PAS is sensitive to CR mutations, related to Figure 7 A. The distribution of PAS from PolyASite 2.0 (Herrmann et al., 2020) around PAS defined from the 3′ end sequencing data. B. The optimal number of k-medoid clusters assessed using the average silhouette method. C. Related to Figure 7A: distribution of the relative change in dPAU (normalized such that WT is always positive) for each gene upon rescue by WT TDP-43 or each variant. Genes have been clustered according to their CR-dependence. D. The ratio of total iCLIP cDNA counts for helix-disrupting (316del346 and A326) normalized against helix-preserving variants (G335A and WT TDP-43) in the region between the proximal and distal PAS for CR-dependent and CR-independent genes. Statistical difference within each group was assessed with a Mann-Whitney test. E. qPCR quantification of the change in cryptic exon usage in the ATG4B gene after expression of TDP-43 variants in combination with siRNA mediated depletion of the endogenous TDP-43 (siT) (n= 3). This cryptic exon has very low expression in Hek-293 cells and this results in qPCR Ct values of 29 versus 25 after TDP-43 depletion.
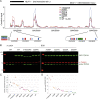
Figure 7
Regulation of a subset of poly(A) sites (PASs) is sensitive to CR mutations (A) The change in usage or delta polyA site usage (dPAU) for the representative PAS in each gene upon rescue by WT TDP-43 or each variant (Table S4). The dPAU for each PAS is normalized so that the WT change is always positive. Red shades indicate rescue in the same direction as the WT and blue shades in the opposite direction; white indicates no rescue. Genes are clustered according to their CR dependence. (B) Top panel: the normalized 316del346 and WT iCLIP coverage tracks (combining all 2,5-HD replicates for each condition) are shown between the proximal (left) and distal (right) PASs that show a regulation pattern that is sensitive to CR mutations (CR-dependent PAS). Center panel: binding motifs in the key binding region (shaded in gray) are magnified and colored by motif type. Bottom panel: the normalized 3′ seq coverage tracks (merging all replicates) for knockdown and rescue with the 316del346 variant and WT. (C) qPCR quantification of the change in PAS usage after expression of TDP-43 variants after knockdown of the endogenous TDP-43 (siT), showing the ratio of the use of distal versus proximal PASs. The shaded region highlights the magnitude of WT rescue over knockdown (n = 3, t test, p = 5.97 × 10−10 [PPP2R2D] and 1.68 × 10−10 [SMC1A]). (D) As in (B) for PASs that are regulated with similar efficiency by all TDP-43 variants regardless of CR mutations (CR-independent PASs). (E) As in (C) for CR-independent PASs. The shaded region highlights the magnitude of WT rescue over knockdown (n = 3, t test, p = 6.97 × 10−8 [GPCPD1] and 0.0158 [GXYLT1]). (F) The ratio of total iCLIP cDNA counts for each TDP-43 variant in the region between the proximal and distal PAS for CR-dependent and CR-independent genes and the ratio of total iCLIP cDNA counts for each TDP-43 variant normalized against WT TDP-43 (combining both biological replicates for each condition). Statistical difference within each group was assessed with an ANOVA. (G) The number of nucleotides covered by TDP-43 bound YG-, YA-, and AA-containing [UG]n motifs in the region between the proximal and distal PASs for CR-dependent and CR-independent genes. Statistical difference was assessed with a Mann-Whitney test. (H) Each RRM domain of TDP-43 recognizes only 4–6 nt in a sequence-specific manner, and TDP-43 binds highly multivalent RNA regions in cells. Condensation-deficient variants of TDP-43 have a decreased capacity to bind a subset of these regions, called “CR-dependent regions”; these tend to be more than 100 nt long and contain a medium density of predominantly YG- and YA-containing [UG]n motifs. The schematic highlights the likely role of CR helix-mediated homomeric interactions in enabling condensation of TDP-43 molecules at a relatively high density on the long CR-dependent regions. Such condensation is regional because long RNAs can contain CR-dependent and CR-independent regions. The conformation of contacts as shown here is purely schematic.
Similar articles
- RNA-binding properties orchestrate TDP-43 homeostasis through condensate formation in vivo.
Scherer NM, Maurel C, Graus MS, McAlary L, Richter G, Radford RAW, Hogan A, Don EK, Lee A, Yerbury J, Francois M, Chung RS, Morsch M. Scherer NM, et al. Nucleic Acids Res. 2024 May 22;52(9):5301-5319. doi: 10.1093/nar/gkae112. Nucleic Acids Res. 2024. PMID: 38381071 Free PMC article. - TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells.
Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A. Colombrita C, et al. J Biol Chem. 2012 May 4;287(19):15635-47. doi: 10.1074/jbc.M111.333450. Epub 2012 Mar 16. J Biol Chem. 2012. PMID: 22427648 Free PMC article. - The implications of physiological biomolecular condensates in amyotrophic lateral sclerosis.
Fakim H, Vande Velde C. Fakim H, et al. Semin Cell Dev Biol. 2024 Mar 15;156:176-189. doi: 10.1016/j.semcdb.2023.05.006. Epub 2023 May 31. Semin Cell Dev Biol. 2024. PMID: 37268555 Review. - TDP-43 in nuclear condensates: where, how, and why.
Lang R, Hodgson RE, Shelkovnikova TA. Lang R, et al. Biochem Soc Trans. 2024 Aug 28;52(4):1809-1825. doi: 10.1042/BST20231447. Biochem Soc Trans. 2024. PMID: 38958608 Free PMC article. Review.
Cited by
- Long way up: rethink diseases in light of phase separation and phase transition.
Ding M, Xu W, Pei G, Li P. Ding M, et al. Protein Cell. 2024 Jul 1;15(7):475-492. doi: 10.1093/procel/pwad057. Protein Cell. 2024. PMID: 38069453 Free PMC article. Review. - Molecular Dissection of TDP-43 as a Leading Cause of ALS/FTLD.
Tamaki Y, Urushitani M. Tamaki Y, et al. Int J Mol Sci. 2022 Oct 19;23(20):12508. doi: 10.3390/ijms232012508. Int J Mol Sci. 2022. PMID: 36293362 Free PMC article. Review. - Pathogenic Mutation of TDP-43 Impairs RNA Processing in a Cell Type-Specific Manner: Implications for the Pathogenesis of ALS/FTLD.
Imaizumi K, Ideno H, Sato T, Morimoto S, Okano H. Imaizumi K, et al. eNeuro. 2022 Jun 8;9(3):ENEURO.0061-22.2022. doi: 10.1523/ENEURO.0061-22.2022. Print 2022 May-Jun. eNeuro. 2022. PMID: 35641224 Free PMC article. - Comparison of TRIBE and STAMP for identifying targets of RNA binding proteins in human and Drosophila cells.
Abruzzi KC, Ratner C, Rosbash M. Abruzzi KC, et al. RNA. 2023 Aug;29(8):1230-1242. doi: 10.1261/rna.079608.123. Epub 2023 May 11. RNA. 2023. PMID: 37169395 Free PMC article. - Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD.
Khalil B, Linsenmeier M, Smith CL, Shorter J, Rossoll W. Khalil B, et al. Mol Neurodegener. 2024 Jan 22;19(1):8. doi: 10.1186/s13024-023-00698-1. Mol Neurodegener. 2024. PMID: 38254150 Free PMC article. Review.
References
- Alberti S., Hyman A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021;22:196–213. - PubMed
- Astoricchio E., Alfano C., Rajendran L., Temussi P.A., Pastore A. The Wide World of Coacervates: From the Sea to Neurodegeneration. Trends Biochem. Sci. 2020;45:706–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- FC001110 /MRC_/Medical Research Council/United Kingdom
- FC001110/WT_/Wellcome Trust/United Kingdom
- FC001110 /CRUK_/Cancer Research UK/United Kingdom
- T32 GM132039/GM/NIGMS NIH HHS/United States
- R01 NS116176/NS/NINDS NIH HHS/United States
- 215593/Z/19/Z /WT_/Wellcome Trust/United Kingdom
- 110292/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 103760/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- 101149/Z/13/A/WT_/Wellcome Trust/United Kingdom
- MR/S006591/1/MRC_/Medical Research Council/United Kingdom
- FC001110 /WT_/Wellcome Trust/United Kingdom
- HALLEGGER/OCT15/959-799/MNDA_/Motor Neurone Disease Association/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous