A network-based systems biology approach for identification of shared Gene signatures between male and female in COVID-19 datasets - PubMed (original) (raw)
A network-based systems biology approach for identification of shared Gene signatures between male and female in COVID-19 datasets
Md Shahjaman et al. Inform Med Unlocked. 2021.
Abstract
The novel coronavirus (SARS-CoV-2) has expanded rapidly worldwide. Now it has covered more than 150 countries worldwide. It is referred to as COVID-19. SARS-CoV-2 mainly affects the respiratory systems of humans that can lead up to serious illness or even death in the presence of different comorbidities. However, most COVID-19 infected people show mild to moderate symptoms, and no medication is suggested. Still, drugs of other diseases have been used to treat COVID-19. Nevertheless, the absence of vaccines and proper drugs against the COVID-19 virus has increased the mortality rate. Albeit sex is a risk factor for COVID-19, none of the studies considered this risk factor for identifying biomarkers from the RNASeq count dataset. Men are more likely to undertake severe symptoms with different comorbidities and show greater mortality compared with women. From this standpoint, we aim to identify shared gene signatures between males and females from the human COVID-19 RNAseq count dataset of peripheral blood cells using a robust voom approach. We identified 1341 overlapping DEGs between male and female datasets. The gene ontology (GO) annotation and pathway enrichment analysis revealed that DEGs are involved in various BP categories such as nucleosome assembly, DNA conformation change, DNA packaging, and different KEGG pathways such as cell cycle, ECM-receptor interaction, progesterone-mediated oocyte maturation, etc. Ten hub-proteins (UBC, KIAA0101, APP, CDK1, SUMO2, SP1, FN1, CDK2, E2F1, and TP53) were unveiled using PPI network analysis. The top three miRNAs (mir-17-5p, mir-20a-5p, mir-93-5p) and TFs (PPARG, E2F1 and KLF5) were uncovered. In conclusion, the top ten significant drugs (roscovitine, curcumin, simvastatin, fulvestrant, troglitazone, alvocidib, L-alanine, tamoxifen, serine, and doxorubicin) were retrieved using drug repurposing analysis of overlapping DEGs, which might be therapeutic agents of COVID-19.
Keywords: COVID-19; Coronavirus; Hub-proteins; Robust voom; SARS-CoV-2; Sex-specific biomarkers.
© 2021 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
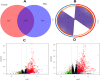
Fig. 1
Differentially expressed genes identification profiles. (A) Venn diagram of DEGs identified by robust voom approach from male and female dataset of COVID-19, (B) circus plot at gene level of overlapping 1341 DEGs between male and female, (C) volcano plot of COVID-19 male dataset, (D) volcano plot of COVID-19 female dataset.
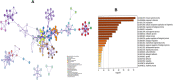
Fig. 2
Heatmap and pathway enrichment analysis of DEGs. (A) KEGG pathway enrichment analysis, (B) heatmap of 10 hub-proteins.
Fig. 3
Gene ontology analysis of DEGs. (A) network of enriched terms colored by cluster identity, (B) barplot of enriched terms using overlapping DEGs colored by p-value.
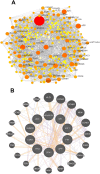
Fig. 4
PPI network analysis of overlapping DEGs. (A) PPI network of 1341 common DEGs identified between male and female, (B) hub-proteins network.
Fig. 5
Performance evaluation of 10 hub-gene using boxplot of Accuracies. (A) boxplot of accuracies of six classifiers, (B) Ranking of 10 hub-genes according to their importance using SVM.
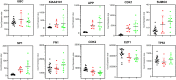
Fig. 6
Gene expression pattern of 10 hub-gene in the control, male and female datasets.
Fig. S1
miRNAs-DEGs regulatory network analysis to identify potential miRNAs
Fig. S2
DEGs-TFs regulatory network analysis to identify potential TFs.
Similar articles
- A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases.
Rahman MH, Peng S, Hu X, Chen C, Rahman MR, Uddin S, Quinn JMW, Moni MA. Rahman MH, et al. Int J Environ Res Public Health. 2020 Feb 6;17(3):1035. doi: 10.3390/ijerph17031035. Int J Environ Res Public Health. 2020. PMID: 32041280 Free PMC article. - Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: Insights from a systems biomedicine perspective.
Rahman MR, Islam T, Zaman T, Shahjaman M, Karim MR, Huq F, Quinn JMW, Holsinger RMD, Gov E, Moni MA. Rahman MR, et al. Genomics. 2020 Mar;112(2):1290-1299. doi: 10.1016/j.ygeno.2019.07.018. Epub 2019 Aug 1. Genomics. 2020. PMID: 31377428 - Identifying molecular insight of synergistic complexities for SARS-CoV-2 infection with pre-existing type 2 diabetes.
Islam MB, Chowdhury UN, Nain Z, Uddin S, Ahmed MB, Moni MA. Islam MB, et al. Comput Biol Med. 2021 Sep;136:104668. doi: 10.1016/j.compbiomed.2021.104668. Epub 2021 Jul 23. Comput Biol Med. 2021. PMID: 34340124 Free PMC article. - Differential microRNA expression in the peripheral blood from human patients with COVID-19.
Li C, Hu X, Li L, Li JH. Li C, et al. J Clin Lab Anal. 2020 Oct;34(10):e23590. doi: 10.1002/jcla.23590. Epub 2020 Sep 22. J Clin Lab Anal. 2020. PMID: 32960473 Free PMC article. - Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods.
Vastrad B, Vastrad C, Tengli A. Vastrad B, et al. 3 Biotech. 2020 Oct;10(10):422. doi: 10.1007/s13205-020-02406-y. Epub 2020 Sep 11. 3 Biotech. 2020. PMID: 33251083 Free PMC article.
Cited by
- Long COVID: Molecular Mechanisms and Detection Techniques.
Constantinescu-Bercu A, Lobiuc A, Căliman-Sturdza OA, Oiţă RC, Iavorschi M, Pavăl NE, Șoldănescu I, Dimian M, Covasa M. Constantinescu-Bercu A, et al. Int J Mol Sci. 2023 Dec 28;25(1):408. doi: 10.3390/ijms25010408. Int J Mol Sci. 2023. PMID: 38203577 Free PMC article. Review. - The SARS-CoV-2 spike S1 protein induces global proteomic changes in ATII-like rat L2 cells that are attenuated by hyaluronan.
Mobley JA, Molyvdas A, Kojima K, Ahmad I, Jilling T, Li JL, Garantziotis S, Matalon S. Mobley JA, et al. Am J Physiol Lung Cell Mol Physiol. 2023 Apr 1;324(4):L413-L432. doi: 10.1152/ajplung.00282.2022. Epub 2023 Jan 31. Am J Physiol Lung Cell Mol Physiol. 2023. PMID: 36719087 Free PMC article. - SARS-CoV-2 potential drugs, drug targets, and biomarkers: a viral-host interaction network-based analysis.
Samy A, Maher MA, Abdelsalam NA, Badr E. Samy A, et al. Sci Rep. 2022 Jul 13;12(1):11934. doi: 10.1038/s41598-022-15898-w. Sci Rep. 2022. PMID: 35831333 Free PMC article. - Discovery of adapalene and dihydrotachysterol as antiviral agents for the Omicron variant of SARS-CoV-2 through computational drug repurposing.
Fidan O, Mujwar S, Kciuk M. Fidan O, et al. Mol Divers. 2023 Feb;27(1):463-475. doi: 10.1007/s11030-022-10440-6. Epub 2022 May 4. Mol Divers. 2023. PMID: 35507211 Free PMC article. - Molecular Mechanisms of Cardiac Injury Associated With Myocardial SARS-CoV-2 Infection.
Liu X, Lou L, Zhou L. Liu X, et al. Front Cardiovasc Med. 2022 Jan 20;8:643958. doi: 10.3389/fcvm.2021.643958. eCollection 2021. Front Cardiovasc Med. 2022. PMID: 35127841 Free PMC article.
References
- Middle East respiratory syndrome coronavirus MERS-CoV. WHO; November 2019. etrieved 20 July 2020.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous