Revised mechanism of hydroxyurea-induced cell cycle arrest and an improved alternative - PubMed (original) (raw)
Revised mechanism of hydroxyurea-induced cell cycle arrest and an improved alternative
Alisa E Shaw et al. Proc Natl Acad Sci U S A. 2024.
Abstract
Replication stress describes endogenous and exogenous challenges to DNA replication in the S-phase. Stress during this critical process causes helicase-polymerase decoupling at replication forks, triggering the S-phase checkpoint, which orchestrates global replication fork stalling and delayed entry into G2. The replication stressor most often used to induce the checkpoint response in yeast is hydroxyurea (HU), a clinically used chemotherapeutic. The primary mechanism of S-phase checkpoint activation by HU has thus far been considered to be a reduction of deoxynucleotide triphosphate synthesis by inhibition of ribonucleotide reductase (RNR), leading to helicase-polymerase decoupling and subsequent activation of the checkpoint, facilitated by the replisome-associated mediator Mrc1. In contrast, we observe that HU causes cell cycle arrest in budding yeast independent of both the Mrc1-mediated replication checkpoint response and the Psk1-Mrc1 oxidative signaling pathway. We demonstrate a direct relationship between HU incubation and reactive oxygen species (ROS) production in yeast and human cells and show that antioxidants restore growth of yeast in HU. We further observe that ROS strongly inhibits the in vitro polymerase activity of replicative polymerases (Pols), Pol α, Pol δ, and Pol ε, causing polymerase complex dissociation and subsequent loss of DNA substrate binding, likely through oxidation of their integral iron-sulfur (Fe-S) clusters. Finally, we present "RNR-deg," a genetically engineered alternative to HU in yeast with greatly increased specificity of RNR inhibition, allowing researchers to achieve fast, nontoxic, and more readily reversible checkpoint activation compared to HU, avoiding harmful ROS generation and associated downstream cellular effects that may confound interpretation of results.
Keywords: ROS; S-phase checkpoint; cell cycle; hydroxyurea; replication stress.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
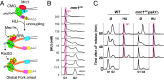
Fig. 1.
HU-induced cell cycle arrest without checkpoint activation or oxidative signaling through Mrc1. (A) Schematic of replication checkpoint. Uncoupling activates Mec1, which phosphorylates Mrc1, activating Rad53, which phosphorylates Mrc1 and CMG, stabilizing forks. (B) Cytometry profiles of _mrc1AQ_are shown at the indicated HU concentration, which was added to asynchronous yeast cells growing in Yeast Peptone Dextrose (YPD) for 4 h prior to flow cytometry acquisition. The red bar indicates G1. (C) Wild Type (WT) or mrc1AQ psk1Δ cells were synced in G1 with αF and released into YPD in the absence or presence of 0.2 M HU, and cytometry profiles were acquired at the indicated times. The red bar indicates G1/S stalling.
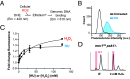
Fig. 2.
Direct relationship between HU incubation and ROS production in yeast cell nuclei. (A) Schematic of DHE assay. H2O2 and other cellular ROS oxidize DHE, converting it to ethidium, which intercalates nuclear DNA and subsequently exhibits red-shifted fluorescence. (B) Cytometry profiles of red-shifted fluorescence intensity from DHE-stained cells in the absence or presence of 0.2 M HU. HU itself does not oxidize DHE (
SI Appendix, Fig. S5
). (C) Dose–response curves of yeast treated with increasing amounts of H2O2 (squares) or HU (circles); nonlinear curve fit R2 values were 0.94 and 0.97, respectively. Error bars represent STD from three replicates. Pearson’s r = 0.85 between both curves. (D) WT or mrc1AQ psk1Δ cells were synced in G1 with αF and released into YPD in the indicated amount of H2O2 and cytometry profiles were acquired after 60 min. The red bar indicates G1/S stalling.
Fig. 3.
ROS inhibits the activity and DNA binding of eukaryotic polymerases by oxidizing their Fe-S clusters and destabilizing the complexes. (A) DNA strand extension activity of Pol α, Pol δ, or Pol ε (which contain Fe-S clusters) or Klenow Pol (which does not) was measured on clamp-loaded, RPA-coated template/primer substrates in the presence of the indicated H2O2 concentrations (Top: assay schematic; Middle: Fe-S clusters are shown in Pri2 for Pol α, the CTD of Pol3 for Pol δ, and the NTD of Pol2 for Pol ε. Bar graphs are quantifications of three separate experiments; error bars represent STD (example gels in
SI Appendix, Fig. S6
). Values above bars (*P < 0.05; ***P < 0.001) indicate a one-tailed equal variance Student’s t test between that concentration and the one preceding it. Where not indicated, the value is not significant. DNA template/primer binding activity of (B) Pol α or (C) Klenow Pol in the presence or absence of 50 mM H2O2 was measured by fluorescence polarization (Top: assay schematic). Mass photometry histograms of (D) Pol α and (E) Pol δ are shown in the presence or absence of 9 mM H2O2. (F) Pol ε is shown in the presence of 45 mM H2O2. Dashed rectangles denote the expected MW of each fully assembled complex. (G) Antioxidants restore growth in HU. Where indicated, cells were plated in media containing 30 mM N-Acetyl Cysteine (NAC) and 90 mM Ascorbic Acid (Asc), with 0.2 M HU where noted. (H) Model of cell cycle arrest by HU independent of RNR inhibition or checkpoint response. HU generates nuclear ROS through a metabolic intermediate, oxidizing integral Fe-S clusters in replicative polymerases, causing polymerase complex dissociation and/or structural changes, halting global replication and stalling the cell cycle (Discussion).
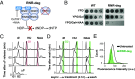
Fig. 4.
RNR-deg is a superior alternative to HU for inducing cell cycle arrest. (A) Schematic of RNR-deg. AID-Rnr1 is degraded by OsTIR1 in the presence of NAA, in an rnr3Δ background, halting dNTP production required for DNA replication. (B) Survival spot assay of WT vs. RNR-deg (OsTIR1, AID-rnr1, rnr3Δ) strains. OsTIR1 expression requires Gal, and degradation requires NAA. Spots contained 5 × 104, 5 × 103, 500, or 50 cells. (C) Flow cytometry profiles of RNR-deg cells that were synchronized in G1 by αF and released into YPG/Gal with the indicated treatment (200 mM HU or 250 µM NAA). (D) RNR-deg treatment reverses faster than HU treatment after washout. Asynchronous RNR-deg cells were treated with αF, released into media containing the indicated treatment for 4 h, and washed extensively by pelleting (1,000×g), then resuspending cells in water two times, followed by a final pelleting/resuspension in YPG/Gal. Cytometry profiles were acquired at the indicated times after washout. Green bar highlights cells in G2. The cell cycle speed of RNR-deg + NAA cells after wash is faster than postwash HU-treated cells, comparable to the speed of untreated cells. (E) Cytometry profiles of red-shifted fluorescence intensity from DHE-stained RNR-deg cells in the absence or presence of 250 µM NAA; the untreated profile from Fig. 2_B_ is used for comparison.
Update of
- Revised Mechanism of Hydroxyurea Induced Cell Cycle Arrest and an Improved Alternative.
Shaw AE, Whitted JE, Mihelich MN, Reitman HJ, Timmerman AJ, Schauer GD. Shaw AE, et al. bioRxiv [Preprint]. 2024 Mar 4:2024.03.02.583010. doi: 10.1101/2024.03.02.583010. bioRxiv. 2024. PMID: 38496404 Free PMC article. Updated. Preprint.
Similar articles
- Revised Mechanism of Hydroxyurea Induced Cell Cycle Arrest and an Improved Alternative.
Shaw AE, Whitted JE, Mihelich MN, Reitman HJ, Timmerman AJ, Schauer GD. Shaw AE, et al. bioRxiv [Preprint]. 2024 Mar 4:2024.03.02.583010. doi: 10.1101/2024.03.02.583010. bioRxiv. 2024. PMID: 38496404 Free PMC article. Updated. Preprint. - A role for Saccharomyces cerevisiae Chk1p in the response to replication blocks.
Schollaert KL, Poisson JM, Searle JS, Schwanekamp JA, Tomlinson CR, Sanchez Y. Schollaert KL, et al. Mol Biol Cell. 2004 Sep;15(9):4051-63. doi: 10.1091/mbc.e03-11-0792. Epub 2004 Jun 30. Mol Biol Cell. 2004. PMID: 15229282 Free PMC article. - In yeast cells arrested at the early S-phase by hydroxyurea, rRNA gene promoters and chromatin are poised for transcription while rRNA synthesis is compromised.
Charton R, Muguet A, Griesenbeck J, Smerdon MJ, Conconi A. Charton R, et al. Mutat Res. 2019 May;815:20-29. doi: 10.1016/j.mrfmmm.2019.04.003. Epub 2019 Apr 27. Mutat Res. 2019. PMID: 31063901 - Hydroxyurea-The Good, the Bad and the Ugly.
Musiałek MW, Rybaczek D. Musiałek MW, et al. Genes (Basel). 2021 Jul 19;12(7):1096. doi: 10.3390/genes12071096. Genes (Basel). 2021. PMID: 34356112 Free PMC article. Review. - The S-phase checkpoint: targeting the replication fork.
Segurado M, Tercero JA. Segurado M, et al. Biol Cell. 2009 Aug 19;101(11):617-27. doi: 10.1042/BC20090053. Biol Cell. 2009. PMID: 19686094 Review.
References
- Reichard P., Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57, 349–374 (1988). - PubMed
MeSH terms
Substances
Grants and funding
- R00 GM126143/GM/NIGMS NIH HHS/United States
- R35 GM147105/GM/NIGMS NIH HHS/United States
- R35GM147105/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R00GM126143/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Miscellaneous