Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and "APS reductase" activity - PubMed (original) (raw)
Comparative Study
Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and "APS reductase" activity
J F Gutierrez-Marcos et al. Proc Natl Acad Sci U S A. 1996.
Abstract
Three different cDNAs, Prh-19, Prh-26, and Prh-43 [3'-phosphoadenosine-5'-phosphosulfate (PAPS) reductase homolog], have been isolated by complementation of an Escherichia coli cysH mutant, defective in PAPS reductase activity, to prototrophy with an Arabidopsis thaliana cDNA library in the expression vector lambda YES. Sequence analysis of the cDNAs revealed continuous open reading frames encoding polypeptides of 465, 458, and 453 amino acids, with calculated molecular masses of 51.3, 50.5, and 50.4 kDa, respectively, that have strong homology with fungal, yeast and bacterial PAPS reductases. However, unlike microbial PAPS reductases, each PRH protein has an N-terminal extension, characteristic of a plastid transit peptide, and a C-terminal extension that has amino acid and deduced three-dimensional homology to thioredoxin proteins. Adenosine 5'-phosphosulfate (APS) was shown to be a much more efficient substrate than PAPS when the activity of the PRH proteins was tested by their ability to convert 35S-labeled substrate to acid-volatile 35S-sulfite. We speculate that the thioredoxin-like domain is involved in catalytic function, and that the PRH proteins may function as novel "APS reductase" enzymes. Southern hybridization analysis showed the presence of a small multigene family in the Arabidopsis genome. RNA blot hybridization with gene-specific probes revealed for each gene the presence of a transcript of approximately 1.85 kb in leaves, stems, and roots that increased on sulfate starvation. To our knowledge, this is the first report of the cloning and characterization of plant genes that encode proteins with APS reductase activity and supports the suggestion that APS can be utilized directly, without activation to PAPS, as an intermediary substrate in reductive sulfate assimilation.
Figures
Figure 1
Comparisons of A. thaliana PRH-19, PRH-26, and PRH-43 deduced amino acid sequences. (A) Sequence comparison with microbial PAPS reductases. Amino acid residues identical to PRH-19 are indicated by a dot. Breaks in the alignment are indicated by a dash. Putative transit peptide cleavage sites in the PRH sequences are underlined. Conserved amino acid regions are indicated by shaded boxes, and other conserved single amino acid residues are indicated by open boxes. Sequences were aligned with the
pileup
program. Compared microbial PAPS reductase sequences are: asper, Aspergillus nidulans SA gene (ref. ; GenBank accession no. X82555X82555); scere, Saccharomyces cerevisiae strain 10197 MET16 gene (ref. ; GenBank accession no. J05591J05591); thioc, Thiocapsa reseopersicina cysH gene (T. Haverkamp, G. Gisselman, and J. D. Schwenn, personal communication; GenBank accession no. Z23169Z23169); ecoli, E. coli K12_cysH_ gene (ref. ; GenBank accession no. Y07525Y07525); styph, Salmonella typhimurium cysH gene (ref. ; GenBank accession no. J05025J05025); anacy, Anacystis nidulans R2 par gene (ref. ; GenBank accession no. M844476). (B) Comparison of the C-terminal sequence of the deduced PRH-19 amino acid sequence with other deduced homologous amino acid sequences. Numbers refer to the amino acid residues in the deduced PRH-19 protein and in the proteins being compared. Conserved amino acid residues are indicated by shaded boxes; highly conserved redox-active cysteine residues are indicated by black boxes. Compared sequences are as follows: PRH19-arab, Arabidopsis PRH19 gene; Pdi-rat, rat protein disulfide isomerase gene (ref. ; GenBank accession no. X02918X02918); ERp72-mouse, mouse endoplasmic reticulum protein gene ERp72 (ref. ; GenBank accession no. J05186J05186); ERp60-bovin, bovine endoplasmic reticulum protein gene ERp60 (Hirano et al., personal communication; GenBank accession no. D16235D16235); ERp5-medsau, golden hamster endoplasmic reticulum protein gene ERp5 (ref. ; GenBank accession no. X62678X62678); ThiM-spiol, spinach thioredoxin_m_ gene (ref. ; GenBank accession no. P07591P07591); ThiF-Pisum, pea thioredoxin f gene (ref. ; GenBank accession no. U35830U35830); ThiH-arab, Arabidopsis thioredoxin h gene (ref. ; GenBank accession no. Z35473Z35473); Grx-oryza, rice glutaredoxin gene (ref. ; GenBank accession no. X77150X77150).
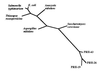
Figure 2
Phylogenetic relationship between PRH proteins and PAPS reductase proteins. Microbial PAPS reductase sequences and the homologous central sequence of PRH-19, PRH-26, and PRH-43 (residues 89–353 with respect to PRH-19) were aligned using
pileup
, and the phylogenetic distances were identified with
phylip
and plotted as a phylogenetic tree.
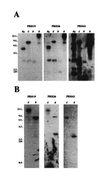
Figure 3
Southern analysis of A. thaliana genomic DNA. (A) Hybridization with either Prh-19, Prh-26, or Prh-43 full-length cDNAs respectively. (B) Hybridization with gene-specific probes derived from the 5′ end of Prh-19, Prh-26, and Prh-43 cDNAs, respectively. Bg, _Bgl_II; E,_Eco_RI; H, _Hin_dIII; B,_Bam_HI.
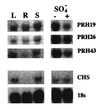
Figure 4
Northern blot analysis of expression of the PRH genes in A. thaliana. (Upper Left) Total leaf (L), root (R), and stem (S) RNA was extracted from 21-day-old compost-grown plants grown under normal sulfur nutrition. (Upper Right) Total RNA was extracted from A. thaliana plants growing in liquid shake culture in MS medium with 1.6 mM sulfate (+ SO4−) or deprived of sulfate for 48h (− SO4−). RNA was hybridized with gene-specific probes derived from the 5′ end of Prh-19, Prh-23, and Prh-43 cDNAs, respectively. (Lower) RNA was hybridized with the chalcone synthase probe (35) and with an 18S rRNA probe (36).
Similar articles
- Sulfate reduction in higher plants: molecular evidence for a novel 5'-adenylylsulfate reductase.
Setya A, Murillo M, Leustek T. Setya A, et al. Proc Natl Acad Sci U S A. 1996 Nov 12;93(23):13383-8. doi: 10.1073/pnas.93.23.13383. Proc Natl Acad Sci U S A. 1996. PMID: 8917600 Free PMC article. - The presence of an iron-sulfur cluster in adenosine 5'-phosphosulfate reductase separates organisms utilizing adenosine 5'-phosphosulfate and phosphoadenosine 5'-phosphosulfate for sulfate assimilation.
Kopriva S, Büchert T, Fritz G, Suter M, Benda R, Schünemann V, Koprivova A, Schürmann P, Trautwein AX, Kroneck PM, Brunold C. Kopriva S, et al. J Biol Chem. 2002 Jun 14;277(24):21786-91. doi: 10.1074/jbc.M202152200. Epub 2002 Apr 8. J Biol Chem. 2002. PMID: 11940598 - Redefining reductive sulfate assimilation in higher plants: a role for APS reductase, a new member of the thioredoxin superfamily?
Wray JL, Campbell EI, Roberts MA, Gutierrez-Marcos JF. Wray JL, et al. Chem Biol Interact. 1998 Feb 20;109(1-3):153-67. doi: 10.1016/s0009-2797(97)00130-0. Chem Biol Interact. 1998. PMID: 9566743 Review. - Identification of a new class of 5'-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria.
Bick JA, Dennis JJ, Zylstra GJ, Nowack J, Leustek T. Bick JA, et al. J Bacteriol. 2000 Jan;182(1):135-42. doi: 10.1128/JB.182.1.135-142.2000. J Bacteriol. 2000. PMID: 10613872 Free PMC article. - The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species.
Klein M, Papenbrock J. Klein M, et al. J Exp Bot. 2004 Aug;55(404):1809-20. doi: 10.1093/jxb/erh183. Epub 2004 Jul 2. J Exp Bot. 2004. PMID: 15234990 Review.
Cited by
- Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism.
Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Maruyama-Nakashita A, et al. Plant Cell. 2006 Nov;18(11):3235-51. doi: 10.1105/tpc.106.046458. Epub 2006 Nov 17. Plant Cell. 2006. PMID: 17114350 Free PMC article. - Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX.
Lee JR, Lee SS, Jang HH, Lee YM, Park JH, Park SC, Moon JC, Park SK, Kim SY, Lee SY, Chae HB, Jung YJ, Kim WY, Shin MR, Cheong GW, Kim MG, Kang KR, Lee KO, Yun DJ, Lee SY. Lee JR, et al. Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5978-83. doi: 10.1073/pnas.0811231106. Epub 2009 Mar 17. Proc Natl Acad Sci U S A. 2009. PMID: 19293385 Free PMC article. - Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome.
Dorrell RG, Gile G, McCallum G, Méheust R, Bapteste EP, Klinger CM, Brillet-Guéguen L, Freeman KD, Richter DJ, Bowler C. Dorrell RG, et al. Elife. 2017 May 12;6:e23717. doi: 10.7554/eLife.23717. Elife. 2017. PMID: 28498102 Free PMC article. - Arabidopsis putative selenium-binding protein1 expression is tightly linked to cellular sulfur demand and can reduce sensitivity to stresses requiring glutathione for tolerance.
Hugouvieux V, Dutilleul C, Jourdain A, Reynaud F, Lopez V, Bourguignon J. Hugouvieux V, et al. Plant Physiol. 2009 Oct;151(2):768-81. doi: 10.1104/pp.109.144808. Epub 2009 Aug 26. Plant Physiol. 2009. PMID: 19710230 Free PMC article. - Sulfur assimilation and the role of sulfur in plant metabolism: a survey.
Droux M. Droux M. Photosynth Res. 2004;79(3):331-48. doi: 10.1023/B:PRES.0000017196.95499.11. Photosynth Res. 2004. PMID: 16328799
References
- Schmidt A, Jäger K. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:325–349.
- Schwenn J D. Z Naturforsch C. 1989;44:504–508.
- Schiffmann S, Schwenn J D. FEBS Lett. 1994;355:229–232. - PubMed
- Murashige T, Skoog F. Physiol Plant. 1962;15:473–497.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous