Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59 - PubMed (original) (raw)
Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59
D L Bodian et al. J Exp Med. 1997.
Abstract
The Ly-6 superfamily of cell surface molecules includes CD59, a potent regulator of the complement system that protects host cells from the cytolytic action of the membrane attack complex (MAC). Although its mechanism of action is not well understood, CD59 is thought to prevent assembly of the MAC by binding to the C8 and/or C9 proteins of the nascent complex. Here a systematic, structure-based mutational approach has been used to determine the region(s) of CD59 required for its protective activity. Analysis of 16 CD59 mutants with single, highly nonconservative substitutions suggests that CD59 has a single active site that includes Trp-40, Arg-53, and Glu-56 of the glycosylated, membrane-distal face of the disk-like extra-cellular domain and, possibly, Asp-24 positioned at the edge of the domain. The putative active site includes residues conserved across species, consistent with the lack of strict homologous restriction previously observed in studies of CD59 function. Competition and mutational analyses of the epitopes of eight CD59-blocking and non-blocking monoclonal antibodies confirmed the location of the active site. Additional experiments showed that the expression and function of CD59 are both glycosylation independent.
Figures
Figure 1
Alignment of CD59 species homologs and HVS-15. Amino acid sequences for the extracellular regions of human (22), African green monkey (11), baboon (11), owl monkey (GenBank accession number: L22861), marmoset (GenBank accession number L22860), squirrel monkey (6), and rat (23) CD59 and Herpesvirus saimiri protein HVS-15 (5) were aligned with the partial sequences of pig (23) and sheep (44) CD59. Residues identical in all sequences are inverse-shaded black and those that are conserved in all species with one exception are inverse-shaded gray. Putative glycosylation sites are boxed. Lower case letters represent probable amino acid assignment, and X represents positions with unknown sequence (23). Residues are numbered according to the sequence of the human mature polypeptide. Every tenth residue is indicated, and both the residue number and introduced substitution are listed for each position selected for mutation. Asterisks mark mutations that disrupted the complement-inhibitory function of human CD59. The sequences were aligned using GCG software (45).
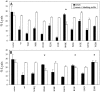
Figure 2
Protection of CD59 mutant-transfected cells from lysis by complement. (A) First-round scanning mutations; (B) second-round mutations. CHO cells transfected with mutant CD59 genes were loaded with calcein-AM and then subjected to complement attack as described in the Materials and Methods. Percent lysis was calculated from the fluorescence of calcein released by cell lysis as a fraction of total calcein loading as also described in Materials and Methods. For all mutants, the total fluorescence per well was similar to the wild-type value. Mean and standard deviations of % lysis in the presence of NHS alone (filled bars) or in the presence of NHS + anti-CD59 blocking antibodies (open bars) were calculated from triplicate samples in a single experiment. Asterisks identify mutants which do not protect cells from lysis. Results are from a single experiment; independent replications gave similar results.
Figure 3
Location and properties of the proposed active site of CD59. (A–C) Mutagenesis data. The residues which were mutated are all numbered and those whose mutation reduced protection (* in Fig. 1) or had no effect are colored red and light blue, respectively. N18, which is N-glycosylated, is colored green. Back (B) and side (C) views differ from the front view (A) by 180° and 90°, respectively. (D) Chemical features. Hydrophobic residues are colored green, polar uncharged residues, light blue, positively charged residues, dark blue, and negatively charged residues, red. The view is the same as in A. (E) Conserved residues. Non-cysteine residues that are conserved in all known CD59 sequences and HVS-15 (inverse-shaded black in Fig. 1) are colored red and those conserved in all sequences with one exception (inverse-shaded gray in Fig. 1) are colored orange. Cysteine residues, which are also conserved in all sequences, are colored yellow. The view is the same as in A. (F) Secondary structure. The positions of the two- and three-stranded β-sheets (purple and red, respectively) and the α-helix (dark blue) of CD59 are shown. Loop residues are colored light blue. The view is the same as in A. All of the experimental data are superimposed on the lowest energy NMR structure (19) drawn using Rasmol (46).
Figure 4
The epitopes of anti-CD59 function-blocking mAb cluster in the region of the proposed active site of CD59. Residues whose mutation disrupted the binding of each antibody (inverse shaded black in Table 1) are shaded black and those whose mutation led to reduced levels of binding (shaded gray in Table 1) are shaded gray. Only antibodies HC1 and MEM43/5 have no CD59-blocking ability. In the last panel, all of the residues that were mutated are shaded gray except for the visible residues whose mutation disrupted the function of CD59 which are shaded black and labeled. In each panel the protein face shown is the one containing the proposed active site shown in the same view as in Fig. 3_A_. The experimental data are superimposed on the lowest energy NMR structure (19) drawn using Rasmol (46).
Figure 5
Western-blot analysis of mutant N18Q. CHO cells transfected with the expression vector alone or with the vector expressing wildtype or mutant N18Q CD59 were solubilized and subjected to SDS-PAGE along with purified erythrocyte CD59 and then Western-blotted with antiCD59 antibody MEM43 as described in the Materials and Methods. Lanes 1, 2, and 3 contained 500, 100, and 50 ng of erythrocyte CD59, respectively. Lanes 4, 5, and 6 contained aliquots of the cell lysate corresponding to ∼5 × 105 CHO cells transfected with the expression vector alone or with the vector expressing mutant N18Q or wild-type CD59, respectively.
Similar articles
- Mapping the active site of CD59.
Yu J, Abagyan R, Dong S, Gilbert A, Nussenzweig V, Tomlinson S. Yu J, et al. J Exp Med. 1997 Feb 17;185(4):745-53. doi: 10.1084/jem.185.4.745. J Exp Med. 1997. PMID: 9034152 Free PMC article. - Elimination of potential sites of glycosylation fails to abrogate complement regulatory function of cell surface CD59.
Rother RP, Zhao J, Zhou Q, Sims PJ. Rother RP, et al. J Biol Chem. 1996 Sep 27;271(39):23842-5. doi: 10.1074/jbc.271.39.23842. J Biol Chem. 1996. PMID: 8798614 - Defining the CD59-C9 binding interaction.
Huang Y, Qiao F, Abagyan R, Hazard S, Tomlinson S. Huang Y, et al. J Biol Chem. 2006 Sep 15;281(37):27398-404. doi: 10.1074/jbc.M603690200. Epub 2006 Jul 14. J Biol Chem. 2006. PMID: 16844690 - Structure, distribution, and functional role of protectin (CD59) in complement-susceptibility and in immunotherapy of human malignancies (Review).
Maio M, Brasoveanu LI, Coral S, Sigalotti L, Lamaj E, Gasparollo A, Visintin A, Altomonte M, Fonsatti E. Maio M, et al. Int J Oncol. 1998 Aug;13(2):305-18. doi: 10.3892/ijo.13.2.305. Int J Oncol. 1998. PMID: 9664126 Review. - CD59: its role in complement regulation and potential for therapeutic use.
Sugita Y, Masuho Y. Sugita Y, et al. Immunotechnology. 1995 Dec;1(3-4):157-68. doi: 10.1016/1380-2933(95)00018-6. Immunotechnology. 1995. PMID: 9373344 Review.
Cited by
- SARS-CoV-2 hijacks host CD55, CD59 and factor H to impair antibody-dependent complement-mediated lysis.
Gebetsberger L, Malekshahi Z, Teutsch A, Tajti G, Fontaine F, Marella N, Mueller A, Prantl L, Stockinger H, Stoiber H, Ohradanova-Repic A. Gebetsberger L, et al. Emerg Microbes Infect. 2024 Dec;13(1):2417868. doi: 10.1080/22221751.2024.2417868. Epub 2024 Oct 28. Emerg Microbes Infect. 2024. PMID: 39435487 Free PMC article. - Structural basis for membrane attack complex inhibition by CD59.
Couves EC, Gardner S, Voisin TB, Bickel JK, Stansfeld PJ, Tate EW, Bubeck D. Couves EC, et al. Nat Commun. 2023 Feb 16;14(1):890. doi: 10.1038/s41467-023-36441-z. Nat Commun. 2023. PMID: 36797260 Free PMC article. - Bispecific mAb2 Antibodies Targeting CD59 Enhance the Complement-Dependent Cytotoxicity Mediated by Rituximab.
Stadlbauer K, Andorfer P, Stadlmayr G, Rüker F, Wozniak-Knopp G. Stadlbauer K, et al. Int J Mol Sci. 2022 May 6;23(9):5208. doi: 10.3390/ijms23095208. Int J Mol Sci. 2022. PMID: 35563599 Free PMC article. - How Structures of Complement Complexes Guide Therapeutic Design.
Bickel JK, Voisin TB, Tate EW, Bubeck D. Bickel JK, et al. Subcell Biochem. 2021;96:273-295. doi: 10.1007/978-3-030-58971-4_7. Subcell Biochem. 2021. PMID: 33252733 Review. - A cryptic non-GPI-anchored cytosolic isoform of CD59 controls insulin exocytosis in pancreatic β-cells by interaction with SNARE proteins.
Golec E, Rosberg R, Zhang E, Renström E, Blom AM, King BC. Golec E, et al. FASEB J. 2019 Nov;33(11):12425-12434. doi: 10.1096/fj.201901007R. Epub 2019 Aug 14. FASEB J. 2019. PMID: 31412214 Free PMC article.
References
- Ryan US. Complement inhibitory therapeutics and xenotransplantation. Nat Med. 1995;1:967–968. - PubMed
- Brooimans R, van Wieringen P, van Es L, Daha MR. Relative roles of decay-accelerating factor, membrane cofactor protein, and CD59 in the protection of human endothelial cells against complement-mediated lysis. Eur J Immunol. 1992;22:3135–3140. - PubMed
- Yamashina M, Ueda E, Kinoshita T, Takami T, Ojima A, Ono H, Tanaka H, Kondo N, Orii T, Okada N, et al. Inherited complete deficiency of 20-kilodalton homologous restriction factor (CD59) as a cause of paroxysmal nocturnal hemoglobinuria. N Engl J Med. 1990;323:1184–1189. - PubMed
- Albrecht J-C, Nicholas J, Cameron KR, Newman C, Fleckenstein B, Honess RW. Herpesvirus Saimiri has a gene specifying a homologue of the cellular membrane glycoprotein CD59. Virology. 1992;190:527–530. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous