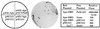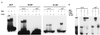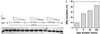Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway - PubMed (original) (raw)
Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway
R A Bennett et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Mutagenic abasic (AP) sites are generated directly by DNA-damaging agents or by DNA glycosylases acting in base excision repair. AP sites are corrected via incision by AP endonucleases, removal of deoxyribose 5-phosphate, repair synthesis, and ligation. Mammalian DNA polymerase beta (Polbeta) carries out most base excision repair synthesis and also can excise deoxyribose 5-phosphate after AP endonuclease incision. Yeast two-hybrid analysis now indicates protein-protein contact between Polbeta and human AP endonuclease (Ape protein). In vitro, binding of Ape protein to uncleaved AP sites loads Polbeta into a ternary complex with Ape and the AP-DNA. After incision by Ape, only Polbeta exhibits stable DNA binding. Kinetic experiments indicated that Ape accelerates the excision of 5'-terminal deoxyribose 5-phosphate by Polbeta. Thus, the two central players of the base excision repair pathway are coordinated in sequential reactions.
Figures
Figure 1
Interaction of Ape and Polβ in the yeast two-hybrid system. (Left) Schematic of plasmids present in indicator strain Y190 (20). Plasmid pAS1–Ape encodes the Gal4DB–Ape fusion; pACT–Polβ encodes the Gal4AD–Polβ fusion; pSE1111 encodes a Gal4AD-Snf4 fusion; pSE1112 encodes a Gal4DB-Snf1 fusion (19). (Center) Indicator plate for β-galactosidase expression from a GAL promoter-lacZ fusion and His+ selection from a GAL promoter-HIS3 fusion. (Right) Quantitation of β-galactosidase expression by a fluorescent assay. Ape-DBD is the Gal4DB–Ape fusion, Polβ-AD the Gal4AD–Polβ fusion. Six independent cotransformants of pAS1-Ape and pACT-Polβ were assayed for β-galactosidase activity; single isolates of the other transformants were assayed. An S. cerevisiae lacZ+ strain (obtained from R. Brennan and R. H. Schiestl, Harvard School of Public Health, Boston) was assayed as a positive control.
Figure 2
Loading of Polβ onto AP sites by Ape protein. (A) Binding to AP sites in different sequence contexts. Purified human Ape protein (0.55 pmol) or Polβ (0.55 pmol) were incubated with the indicated 5′-labeled AP substrates in reactions containing either 4 mM EDTA or 10 mM MgCl2 (Mg2+), and the uncomplexed DNA (lowest bands) and protein DNA complexes (upper bands) resolved by electrophoresis in a nondenaturing gel (22). Binding substrates were 23-F, a 23-bp duplex DNA containing a tetrahydrofuran residue (10); 18-AP, an 18-bp oligonucleotide containing an AP site generated by uracil excision (10); and 51-AP, a 51-bp duplex oligonucleotide containing an AP site generated by uracil excision (13). (B) Presence of Ape and Polβ in complexes. Binding reactions with EDTA or Mg2+ were carried out with Ape or Polβ and the 51-AP substrate as described above, then Ape-specific (αApe) or Polβ-specific (αβ-pol) antisera were added. The complexes containing Ape or Polβ (middle bands) were resolved from the antibody-supershifted complexes (top bands) by electrophoresis in nondenaturing gels. In no case was significant material retained in the wells of the gels.
Figure 3
Activation of Polβ dRp excision activity by Ape. (A) A duplex oligonucleotide containing an AP site at position 22 was cleaved with a catalytic amount of E. coli endonuclease IV, then incubated for the indicated times with purified human Polβ (9 fmol) and varying amounts of purified human Ape protein (0, 8, 80, or 800 fmol). After the incubation, the substrate bearing 5′-dRp (upper band) and the product after dRp excision (lower band) were resolved in a denaturing gel. OH−, substrate hydrolyzed with NaOH; X, the 5′-dRp substrate before incubation; 15, 5′-dRp substrate incubated 15 min with 800 fmol of Ape alone. (B) Quantitation of dRp excision. The gel in A was subjected to scanning densitometry and the ratio of the substrate (upper band in A) to product (lower band in A) used to calculate the excision of dRp in 25-min reactions.
Similar articles
- Reconstitution of the DNA base excision-repair pathway.
Dianov G, Lindahl T. Dianov G, et al. Curr Biol. 1994 Dec 1;4(12):1069-76. doi: 10.1016/s0960-9822(00)00245-1. Curr Biol. 1994. PMID: 7535646 - Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases.
Erzberger JP, Barsky D, Schärer OD, Colvin ME, Wilson DM 3rd. Erzberger JP, et al. Nucleic Acids Res. 1998 Jun 1;26(11):2771-8. doi: 10.1093/nar/26.11.2771. Nucleic Acids Res. 1998. PMID: 9592167 Free PMC article. - Regulation of eukaryotic abasic endonucleases and their role in genetic stability.
Demple B, Harrison L, Wilson DM 3rd, Bennett RA, Takagi T, Ascione AG. Demple B, et al. Environ Health Perspect. 1997 Jun;105 Suppl 4(Suppl 4):931-4. doi: 10.1289/ehp.97105s4931. Environ Health Perspect. 1997. PMID: 9255583 Free PMC article. Review. - Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1).
Klungland A, Lindahl T. Klungland A, et al. EMBO J. 1997 Jun 2;16(11):3341-8. doi: 10.1093/emboj/16.11.3341. EMBO J. 1997. PMID: 9214649 Free PMC article. - When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions.
Quiñones JL, Demple B. Quiñones JL, et al. DNA Repair (Amst). 2016 Aug;44:103-109. doi: 10.1016/j.dnarep.2016.05.014. Epub 2016 May 20. DNA Repair (Amst). 2016. PMID: 27264558 Free PMC article. Review.
Cited by
- Changes in organization of Crithidia fasciculata kinetoplast DNA replication proteins during the cell cycle.
Johnson CE, Englund PT. Johnson CE, et al. J Cell Biol. 1998 Nov 16;143(4):911-9. doi: 10.1083/jcb.143.4.911. J Cell Biol. 1998. PMID: 9817750 Free PMC article. - Base excision repair of oxidative DNA damage and association with cancer and aging.
Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Maynard S, et al. Carcinogenesis. 2009 Jan;30(1):2-10. doi: 10.1093/carcin/bgn250. Epub 2008 Oct 31. Carcinogenesis. 2009. PMID: 18978338 Free PMC article. Review. - Association between polymorphisms of APE1 and OGG1 and risk of colorectal cancer in Taiwan.
Lai CY, Hsieh LL, Tang R, Santella RM, Chang-Chieh CR, Yeh CC. Lai CY, et al. World J Gastroenterol. 2016 Mar 28;22(12):3372-80. doi: 10.3748/wjg.v22.i12.3372. World J Gastroenterol. 2016. PMID: 27022219 Free PMC article. - PARP1 recruits DNA translocases to restrain DNA replication and facilitate DNA repair.
Ho YC, Ku CS, Tsai SS, Shiu JL, Jiang YZ, Miriam HE, Zhang HW, Chen YT, Chiu WT, Chang SB, Shen CH, Myung K, Chi P, Liaw H. Ho YC, et al. PLoS Genet. 2022 Dec 13;18(12):e1010545. doi: 10.1371/journal.pgen.1010545. eCollection 2022 Dec. PLoS Genet. 2022. PMID: 36512630 Free PMC article.
References
- Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995.
- Lindahl T. Nature (London) 1993;362:709–715. - PubMed
- Wood R D. Annu Rev Biochem. 1996;65:135–167. - PubMed
- Sancar A. Annu Rev Biochem. 1996;65:43–81. - PubMed
- Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA62845/CA/NCI NIH HHS/United States
- R01 GM040000/GM/NIGMS NIH HHS/United States
- T32 CA009078/CA/NCI NIH HHS/United States
- F32 CA062845/CA/NCI NIH HHS/United States
- T32 ES007155/ES/NIEHS NIH HHS/United States
- ES07155/ES/NIEHS NIH HHS/United States
- CA09078/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous