Activities and response to DNA damage of latent and active sequence-specific DNA binding forms of mouse p53 - PubMed (original) (raw)
Activities and response to DNA damage of latent and active sequence-specific DNA binding forms of mouse p53
Y Wu et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The mouse p53 protein generated by alternative splicing (p53as) has amino acid substitutions at its C terminus that result in constitutively active sequence-specific DNA binding (active form), whereas p53 protein itself binds inefficiently (latent form) unless activated by C-terminal modification. Exogenous p53as expression activated transcription of reporter plasmids containing p53 binding sequences and inhibited growth of mouse and human cells lacking functional endogenous p53. Inducible p53as in stably transfected p53 null fibroblasts increased p21(WAF1/Cip-1/Sdi) and decreased bcl-2 protein steady-state levels. Endogenous p53as and p53 proteins differed in response to cellular DNA damage. p53 protein was induced transiently in normal keratinocytes and fibroblasts whereas p53as protein accumulation was sustained in parallel with induction of p21(WAF1/Cip-1/Sdi) protein and mRNA, in support of p53as transcriptional activity. Endogenous p53 and p53as proteins in epidermal tumor cells responded to DNA damage with different kinetics of nuclear accumulation and efficiencies of binding to a p53 consensus DNA sequence. A model is proposed in which C-terminally distinct p53 protein forms specialize in functions, with latent p53 forms primarily for rapid non-sequence-specific binding to sites of DNA damage and active p53 forms for sustained regulation of transcription and growth.
Figures
Figure 1
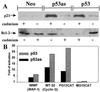
Growth inhibition assays. (A) Saos2 cells were transfected with plasmids containing a neo resistance gene and either no insert, p53, or p53as cDNA. After selection in G418 for 3 weeks, fixed cells were stained with Giemsa. Mean total colony area (mm2) per four 60-mm dishes is shown. The data were consistent in two experiments. (B) Stable (10)1 fibroblast clones containing neo resistance control plasmid, alone or cotransfected with plasmids containing inducible p53as (representative of two clones with integrated p53as), or p53 (four clones), were exposed to CdCl2 (0 hr), and relative cell number was determined as described in Materials and Methods.
Figure 2
Effects on p53 downstream genes in (10)1 cells. (A) Stable inducible p53, p53as, or neo control cell clones were exposed to CdCl2 as in Fig. 1_B_. After 18 hr cells were lysed and 50 μg of protein/lane was immunoblotted using anti-p21 or anti-bcl-2 antibodies. (B) Transactivation of reporter plasmids transiently cotransfected with pCMVp53as, pCMVp53, or pCMV. Cells were lysed after 40–48 hr, and chloramphenicol acetyl transferase (PG13, MG15) or luciferase activity (WWP, WT-30) was measured, standardized to total protein, and expressed relative to pCMV control as 1. Fold activation in three replicate experiments was 3–20 (WWP), 5–18 (WT-30), and 9–15 (PG13).
Figure 3
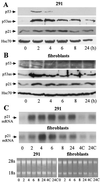
Response to x-ray (400 rads) of normal (A) 291 epidermal cells or secondary mouse fibroblasts (B) detectable by Western immunoblotting. Proteins were extracted at the indicated times postirradiation and reacted with PAb122 (p53), ApAs (p53as), mp21 (p21), or anti-Hsc70 as loading control. (C) Northern blots. Cells were lysed and 7 μg total RNA per lane was separated in 1.2% formaldehyde agarose, transferred to a membrane, and probed using a radiolabeled fragment of mouse p21/waf1 cDNA. Controls included ethidium bromide staining of rRNA (for loading) and cells exposed to solvent for 4 (4C) or 24 hr (24C).
Figure 4
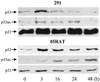
Response to 60 μM genistein detectable by immunoblotting. 291 cells and derivative tumor cells 291.05RAT (05RAT) were treated for the times indicated, and proteins were extracted and analyzed as in Fig. 3.
Figure 5
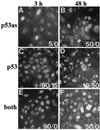
p53as and p53 detection by indirect immunofluorescence. 291.05RAT cells were treated with 60 μM genistein for the times indicated, fixed, and immunostained with ApAs (p53as), PAb421 (p53), 6.2 (both), or IgG/preimmune controls (negative, i.e., <1 staining intensity on scale of 1 to 4; data not shown). Positive cells (intensity, 2.5–4) per total cells per coverslip (%) for nuclear/cytoplasmic staining are shown (at 0 hr, 0.1–5% reacted with antibodies). The peak nuclear antibody reactivity for PAb421 (3 hr) and for ApAs (24–48 hr) is representative of six experiments.
Figure 6
Electrophoretic mobility-shift assay. 291.05RAT cells were harvested at the times indicated after treatment with 60 μM genistein. Equal amounts of protein were reacted with a 32P-labeled p53 consensus DNA sequence in the presence or absence of 200 ng of the antibodies indicated and visualized by autoradiography. The positions of migration of baculovirus-purified recombinant human p53 + PAb421 (solid arrow) and mouse p53as (open arrow) are shown. Free probe migrated at twice the distance of the rapidly migrating (unlabeled) nonspecific band and was present in vast excess of the signals shown.
Figure 7
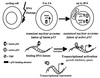
Model: p53 forms respond to DNA damage with distinct kinetics. The p53 forms latent for sequence-specific DNA binding are induced rapidly and specialize in nonspecific DNA binding at sites of DNA damage. Transcription is inhibited through catalysis of strand annealing or by binding to TATA box binding protein (TBP). Forms of p53 active for sequence-specific DNA binding are sustained to evoke long-lasting effects on p53 downstream gene transcription, growth arrest, apoptosis, or cell differentiation. Latent p53 may be activated through posttranslational modifications or binding to single-stranded DNA by-products of nucleotide excision repair, targeting sequence-specific transcriptional effects to sites of intact DNA.
Similar articles
- Repression of transcription and interference with DNA binding of TATA-binding protein by C-terminal alternatively spliced p53.
Huang H, Kaku S, Knights C, Park B, Clifford J, Kulesz-Martin M. Huang H, et al. Exp Cell Res. 2002 Oct 1;279(2):248-59. doi: 10.1006/excr.2002.5596. Exp Cell Res. 2002. PMID: 12243750 - DNA binding specificity of proteins derived from alternatively spliced mouse p53 mRNAs.
Miner Z, Kulesz-Martin M. Miner Z, et al. Nucleic Acids Res. 1997 Apr 1;25(7):1319-26. doi: 10.1093/nar/25.7.1319. Nucleic Acids Res. 1997. PMID: 9060424 Free PMC article. - Wild-type alternatively spliced p53: binding to DNA and interaction with the major p53 protein in vitro and in cells.
Wu Y, Liu Y, Lee L, Miner Z, Kulesz-Martin M. Wu Y, et al. EMBO J. 1994 Oct 17;13(20):4823-30. doi: 10.1002/j.1460-2075.1994.tb06808.x. EMBO J. 1994. PMID: 7957051 Free PMC article. - The murine C'-terminally alternatively spliced form of p53 induces attenuated apoptosis in myeloid cells.
Almog N, Li R, Peled A, Schwartz D, Wolkowicz R, Goldfinger N, Pei H, Rotter V. Almog N, et al. Mol Cell Biol. 1997 Feb;17(2):713-22. doi: 10.1128/MCB.17.2.713. Mol Cell Biol. 1997. PMID: 9001225 Free PMC article. - Signaling to p53: breaking the posttranslational modification code.
Appella E, Anderson CW. Appella E, et al. Pathol Biol (Paris). 2000 Apr;48(3):227-45. Pathol Biol (Paris). 2000. PMID: 10858956 Review.
Cited by
- p53-regulated apoptosis is differentiation dependent in ultraviolet B-irradiated mouse keratinocytes.
Tron VA, Trotter MJ, Tang L, Krajewska M, Reed JC, Ho VC, Li G. Tron VA, et al. Am J Pathol. 1998 Aug;153(2):579-85. doi: 10.1016/S0002-9440(10)65600-3. Am J Pathol. 1998. PMID: 9708817 Free PMC article. - Species-specific regulation of alternative splicing in the C-terminal region of the p53 tumor suppressor gene.
Laverdière M, Beaudoin J, Lavigueur A. Laverdière M, et al. Nucleic Acids Res. 2000 Mar 15;28(6):1489-97. doi: 10.1093/nar/28.6.1489. Nucleic Acids Res. 2000. PMID: 10684946 Free PMC article. - Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation.
Kapoor M, Lozano G. Kapoor M, et al. Proc Natl Acad Sci U S A. 1998 Mar 17;95(6):2834-7. doi: 10.1073/pnas.95.6.2834. Proc Natl Acad Sci U S A. 1998. PMID: 9501176 Free PMC article. - Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans.
Lee BP, Pilling LC, Emond F, Flurkey K, Harrison DE, Yuan R, Peters LL, Kuchel GA, Ferrucci L, Melzer D, Harries LW. Lee BP, et al. Aging Cell. 2016 Oct;15(5):903-13. doi: 10.1111/acel.12499. Epub 2016 Jun 30. Aging Cell. 2016. PMID: 27363602 Free PMC article. - p53-mediated regulation of proliferating cell nuclear antigen expression in cells exposed to ionizing radiation.
Xu J, Morris GF. Xu J, et al. Mol Cell Biol. 1999 Jan;19(1):12-20. doi: 10.1128/MCB.19.1.12. Mol Cell Biol. 1999. PMID: 9858527 Free PMC article.
References
- Pavletich N P, Chambers K A, Pabo C O. Genes Dev. 1993;7:2556–2564. - PubMed
- Cho Y, Gorina S, Jeffery P D, Pavletich N P. Science. 1994;265:346–355. - PubMed
- Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. Genes Dev. 1993;7:2565–2574. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous